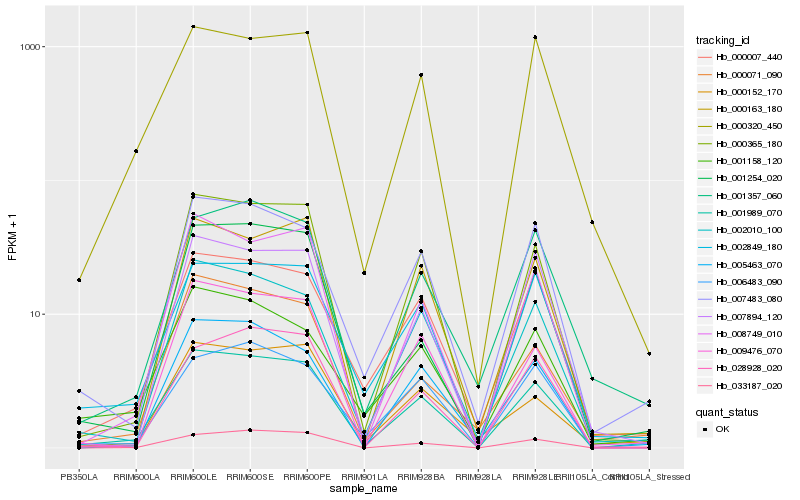
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001989_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized calcium-binding protein At1g02270 [Jatropha curcas] |
| 2 |
Hb_000365_180 |
0.0790595532 |
- |
- |
PREDICTED: protein LURP-one-related 8-like [Pyrus x bretschneideri] |
| 3 |
Hb_002010_100 |
0.1002727625 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 4 |
Hb_008749_010 |
0.1028473801 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 5 |
Hb_007483_080 |
0.1054124305 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 6 |
Hb_002849_180 |
0.1153554454 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 7 |
Hb_033187_020 |
0.1154034247 |
- |
- |
hypothetical protein CARUB_v10004250mg, partial [Capsella rubella] |
| 8 |
Hb_005463_070 |
0.1184027444 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_000007_440 |
0.1185940419 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 10 |
Hb_009476_070 |
0.1194778672 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 11 |
Hb_000163_180 |
0.1227896527 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 2 [Jatropha curcas] |
| 12 |
Hb_001254_020 |
0.1239779067 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 13 |
Hb_007894_120 |
0.1259131103 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001357_060 |
0.1275779567 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 15 |
Hb_001158_120 |
0.1290403298 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 16 |
Hb_028928_020 |
0.1316849565 |
- |
- |
hypothetical protein CICLE_v10018504mg [Citrus clementina] |
| 17 |
Hb_000152_170 |
0.132077652 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 18 |
Hb_006483_090 |
0.1332097803 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 19 |
Hb_000320_450 |
0.1337360815 |
- |
- |
secretory peroxidase [Hevea brasiliensis] |
| 20 |
Hb_000071_090 |
0.1344121088 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |