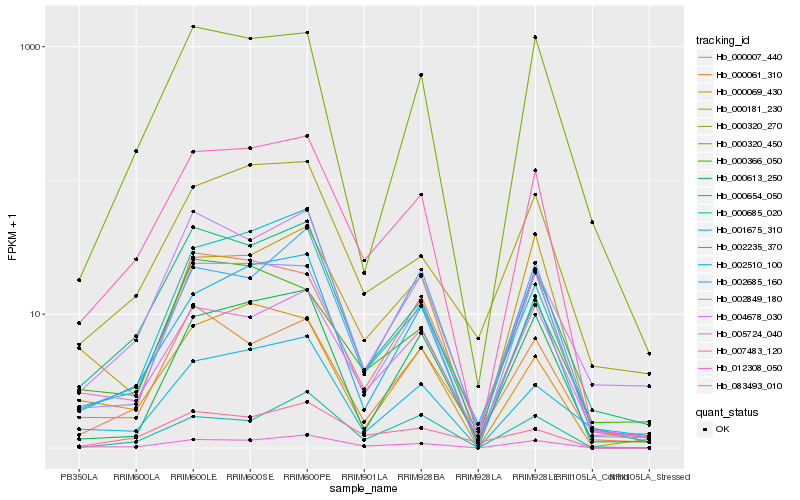
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012308_050 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 2 |
Hb_007483_120 |
0.081404985 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 3 |
Hb_002510_100 |
0.1013528179 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 4 |
Hb_000654_050 |
0.103133365 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 5 |
Hb_002849_180 |
0.106679199 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 6 |
Hb_005724_040 |
0.1265562197 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 7 |
Hb_000320_270 |
0.1285476452 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000320_450 |
0.1410082078 |
- |
- |
secretory peroxidase [Hevea brasiliensis] |
| 9 |
Hb_001675_310 |
0.1424561453 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000613_250 |
0.1433325326 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 11 |
Hb_004678_030 |
0.1441031421 |
- |
- |
fructokinase [Manihot esculenta] |
| 12 |
Hb_000069_430 |
0.1448967038 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 13 |
Hb_000181_230 |
0.1465252461 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_000007_440 |
0.1479552757 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 15 |
Hb_002685_160 |
0.150773755 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 16 |
Hb_002235_370 |
0.1525388685 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 17 |
Hb_083493_010 |
0.1536737736 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 18 |
Hb_000685_020 |
0.1548760999 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 19 |
Hb_000061_310 |
0.1552463966 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000366_050 |
0.1553980108 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |