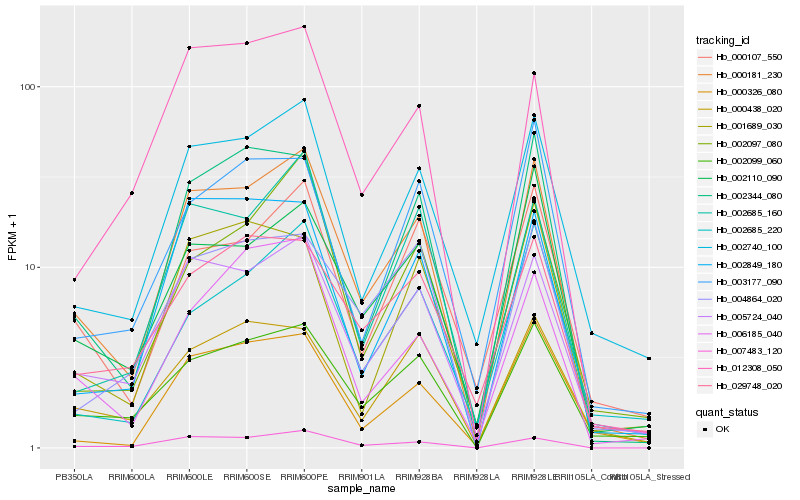
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_230 |
0.0 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_005724_040 |
0.0873595282 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 3 |
Hb_007483_120 |
0.1029870336 |
- |
- |
60S ribosomal protein L32A [Hevea brasiliensis] |
| 4 |
Hb_002740_100 |
0.1063231899 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_002099_060 |
0.1131586378 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 6 |
Hb_002344_080 |
0.1138599056 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 7 |
Hb_002685_160 |
0.1235079779 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 8 |
Hb_002110_090 |
0.1290075382 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 9 |
Hb_000107_550 |
0.132339205 |
- |
- |
PREDICTED: monosaccharide-sensing protein 2 [Jatropha curcas] |
| 10 |
Hb_002849_180 |
0.1363547048 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 11 |
Hb_000326_080 |
0.1377884638 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_029748_020 |
0.1423464433 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_004864_020 |
0.1430371832 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 14 |
Hb_006185_040 |
0.1434867708 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 15 |
Hb_000438_020 |
0.1438984811 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 16 |
Hb_012308_050 |
0.1465252461 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 17 |
Hb_002685_220 |
0.1467603077 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_003177_090 |
0.149218092 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 19 |
Hb_002097_080 |
0.149729921 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 20 |
Hb_001689_030 |
0.1501963232 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |