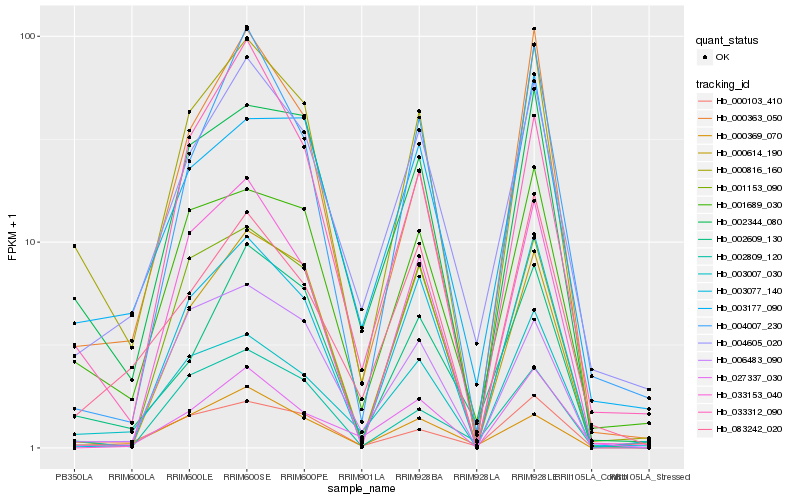
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_160 |
0.0 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 2 |
Hb_002344_080 |
0.1008391168 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 3 |
Hb_001689_030 |
0.1134471019 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 4 |
Hb_000363_050 |
0.128600989 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_003077_140 |
0.1288634854 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 6 |
Hb_003007_030 |
0.1310253976 |
- |
- |
- |
| 7 |
Hb_004605_020 |
0.1341555455 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 8 |
Hb_000103_410 |
0.1346638859 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 9 |
Hb_002609_130 |
0.1395919977 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 10 |
Hb_001153_090 |
0.1434305188 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 11 |
Hb_000614_190 |
0.1449054524 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 12 |
Hb_002809_120 |
0.1452424963 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 13 |
Hb_027337_030 |
0.1469631642 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 14 |
Hb_033153_040 |
0.147173489 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_003177_090 |
0.1519204984 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 16 |
Hb_004007_230 |
0.1520942702 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 17 |
Hb_083242_020 |
0.1521483665 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 18 |
Hb_006483_090 |
0.1545513045 |
- |
- |
PREDICTED: uncharacterized protein LOC105639979 [Jatropha curcas] |
| 19 |
Hb_033312_090 |
0.1549354833 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000369_070 |
0.1559699295 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |