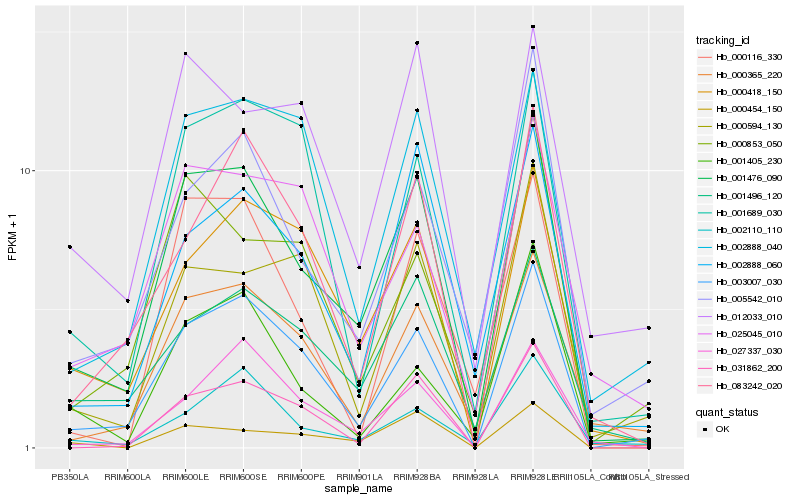
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001476_090 |
0.0 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 2 |
Hb_003007_030 |
0.1083972195 |
- |
- |
- |
| 3 |
Hb_001496_120 |
0.1401187819 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000365_220 |
0.1463192376 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 5 |
Hb_000418_150 |
0.1465328747 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_005542_010 |
0.1525787579 |
- |
- |
Uncharacterized protein TCM_021447 [Theobroma cacao] |
| 7 |
Hb_002888_060 |
0.154875579 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000454_150 |
0.1566760349 |
transcription factor |
TF Family: C3H |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 9 |
Hb_002888_040 |
0.1620805163 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 10 |
Hb_083242_020 |
0.1626924467 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |
| 11 |
Hb_012033_010 |
0.1654598908 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 12 |
Hb_031862_200 |
0.1668120307 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 13 |
Hb_027337_030 |
0.1672067848 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 14 |
Hb_001689_030 |
0.1676803182 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 15 |
Hb_002110_110 |
0.1697731076 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 16 |
Hb_025045_010 |
0.1698219443 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_001405_230 |
0.1719741342 |
- |
- |
STELAR K+ outward rectifier isoform 2 [Theobroma cacao] |
| 18 |
Hb_000853_050 |
0.1720464296 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000594_130 |
0.1722801878 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_000116_330 |
0.1734671475 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |