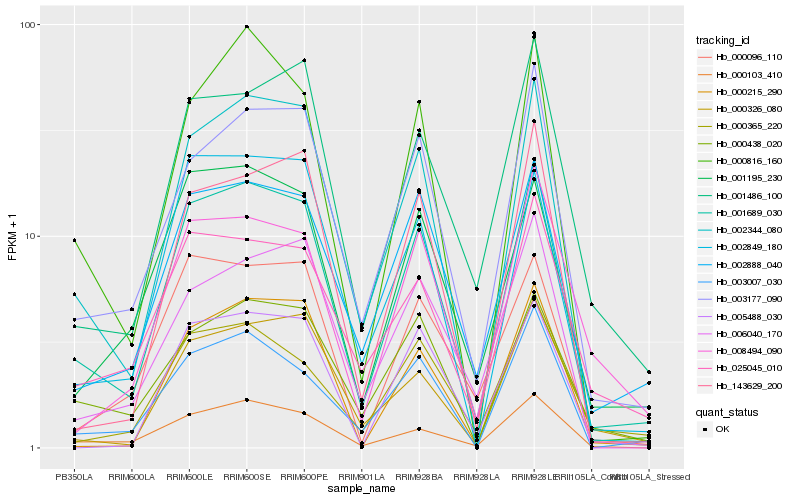
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001689_030 |
0.0 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 2 |
Hb_002344_080 |
0.0793072091 |
- |
- |
PREDICTED: protein YLS2-like [Populus euphratica] |
| 3 |
Hb_003177_090 |
0.0960683651 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 4 |
Hb_000103_410 |
0.1010575615 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 5 |
Hb_002888_040 |
0.1048783117 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 6 |
Hb_000816_160 |
0.1134471019 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 7 |
Hb_003007_030 |
0.1150286034 |
- |
- |
- |
| 8 |
Hb_001486_100 |
0.1187958701 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 9 |
Hb_000438_020 |
0.12040396 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 10 |
Hb_001195_230 |
0.1230536228 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 11 |
Hb_025045_010 |
0.1239985715 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_143629_200 |
0.1249104699 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 13 |
Hb_000326_080 |
0.128201442 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 14 |
Hb_002849_180 |
0.1286721688 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 15 |
Hb_000365_220 |
0.1313416217 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 16 |
Hb_000215_290 |
0.1313596691 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000096_110 |
0.1333769294 |
- |
- |
PREDICTED: uncharacterized protein LOC105787685 isoform X1 [Gossypium raimondii] |
| 18 |
Hb_008494_090 |
0.1347703495 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 19 |
Hb_006040_170 |
0.1353344792 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 20 |
Hb_005488_030 |
0.1354945471 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |