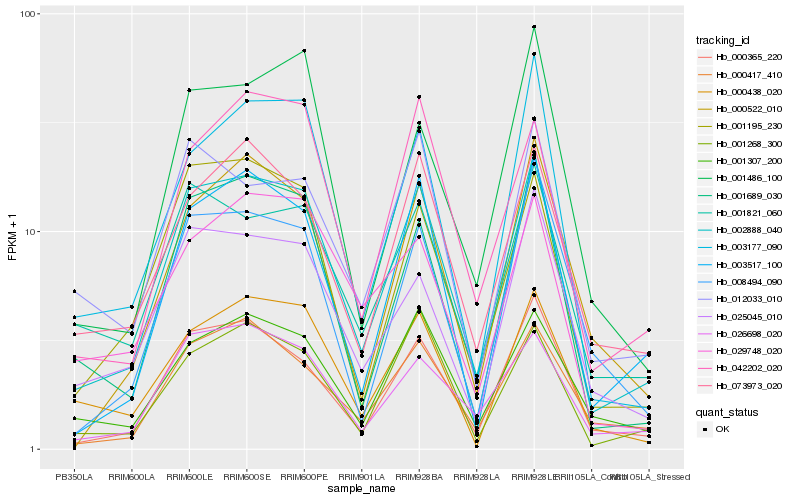
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002888_040 |
0.0 |
- |
- |
hypothetical protein B456_009G267200 [Gossypium raimondii] |
| 2 |
Hb_000365_220 |
0.0939060546 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 3 |
Hb_026698_020 |
0.1034624547 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 4 |
Hb_001689_030 |
0.1048783117 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 5 |
Hb_003517_100 |
0.1060683082 |
transcription factor |
TF Family: GRAS |
- |
| 6 |
Hb_001195_230 |
0.1093882996 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 7 |
Hb_001268_300 |
0.1123167102 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001307_200 |
0.1126760609 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 9 |
Hb_073973_020 |
0.1136924408 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 10 |
Hb_000417_410 |
0.1142814021 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001821_060 |
0.1158246373 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 12 |
Hb_042202_020 |
0.1169246404 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 13 |
Hb_003177_090 |
0.1183089067 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 14 |
Hb_025045_010 |
0.1183565788 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000438_020 |
0.1209920692 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 16 |
Hb_000522_010 |
0.1269669313 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 1-like [Jatropha curcas] |
| 17 |
Hb_008494_090 |
0.1303036699 |
- |
- |
PREDICTED: uncharacterized protein LOC105636007 [Jatropha curcas] |
| 18 |
Hb_012033_010 |
0.1311822251 |
- |
- |
PREDICTED: histone H1.2-like [Jatropha curcas] |
| 19 |
Hb_029748_020 |
0.13393095 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001486_100 |
0.1341707511 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |