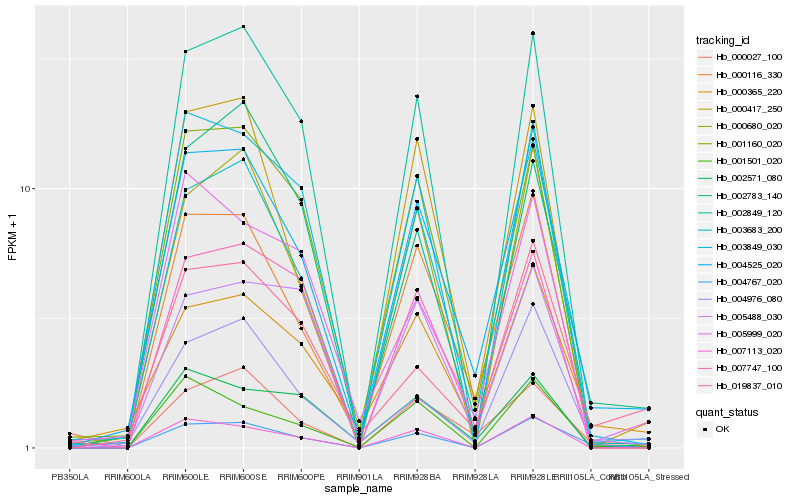
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004525_020 |
0.0 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 2 |
Hb_000116_330 |
0.0967690066 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 3 |
Hb_000027_100 |
0.1048824611 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 4 |
Hb_003683_200 |
0.1064964907 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 5 |
Hb_000365_220 |
0.1245674881 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 6 |
Hb_001160_020 |
0.1247149538 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 7 |
Hb_000417_250 |
0.125573845 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 8 |
Hb_003849_030 |
0.1318168828 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 9 |
Hb_004976_080 |
0.1319266042 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 10 |
Hb_002849_120 |
0.1340739333 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 11 |
Hb_000680_020 |
0.1395803395 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_004767_020 |
0.1423908643 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_002783_140 |
0.1477291154 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 14 |
Hb_019837_010 |
0.1486959113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007113_020 |
0.149670078 |
- |
- |
- |
| 16 |
Hb_002571_080 |
0.1497788938 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |
| 17 |
Hb_007747_100 |
0.1514538819 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 18 |
Hb_001501_020 |
0.1521070427 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 19 |
Hb_005999_020 |
0.1522134592 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 20 |
Hb_005488_030 |
0.1527388024 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |