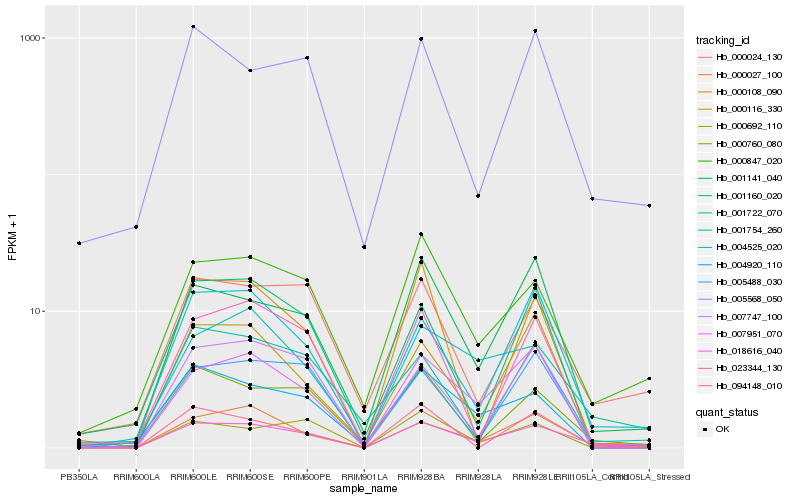
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018616_040 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000027_100 |
0.150029086 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 3 |
Hb_007951_070 |
0.1599060479 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 4 |
Hb_004525_020 |
0.1699590816 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 5 |
Hb_004920_110 |
0.1749927537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000847_020 |
0.1776878116 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 7 |
Hb_000760_080 |
0.1798399931 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 8 |
Hb_005488_030 |
0.1818885314 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 9 |
Hb_001141_040 |
0.1819610396 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 10 |
Hb_023344_130 |
0.1824271439 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_001754_260 |
0.1839524121 |
- |
- |
PREDICTED: uncharacterized protein LOC105643409 isoform X1 [Jatropha curcas] |
| 12 |
Hb_094148_010 |
0.1855699407 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase protein kinase domain-containing GDPDL2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000116_330 |
0.1902448122 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 14 |
Hb_000692_110 |
0.1902597888 |
- |
- |
PREDICTED: NEP1-interacting protein-like 1 [Jatropha curcas] |
| 15 |
Hb_001160_020 |
0.1903881513 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 16 |
Hb_007747_100 |
0.1911539332 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 17 |
Hb_005568_050 |
0.1917070038 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 18 |
Hb_000024_130 |
0.1942825039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001722_070 |
0.1950058877 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 20 |
Hb_000108_090 |
0.1952188864 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |