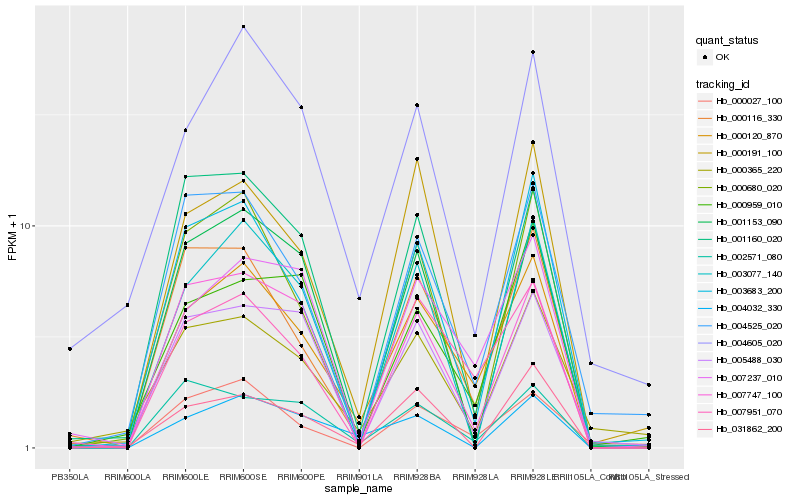
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_870 |
0.0 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 2 |
Hb_007951_070 |
0.1029695268 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_007237_010 |
0.1353558494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000116_330 |
0.1435215642 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 5 |
Hb_003683_200 |
0.1517261551 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 6 |
Hb_000191_100 |
0.1587262446 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_003077_140 |
0.1631211412 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 8 |
Hb_004525_020 |
0.1636445373 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 9 |
Hb_000027_100 |
0.1640544144 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 10 |
Hb_007747_100 |
0.1673191024 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 11 |
Hb_001160_020 |
0.1680181462 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 12 |
Hb_001153_090 |
0.172739221 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 13 |
Hb_004605_020 |
0.1751006677 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 14 |
Hb_005488_030 |
0.1757461919 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_004032_330 |
0.1760956811 |
- |
- |
PREDICTED: calvin cycle protein CP12-3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_031862_200 |
0.1761951859 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Vitis vinifera] |
| 17 |
Hb_000959_010 |
0.1764288713 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000365_220 |
0.1774151187 |
- |
- |
protein phosphatase-1, putative [Ricinus communis] |
| 19 |
Hb_000680_020 |
0.179004712 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_002571_080 |
0.1795244446 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |