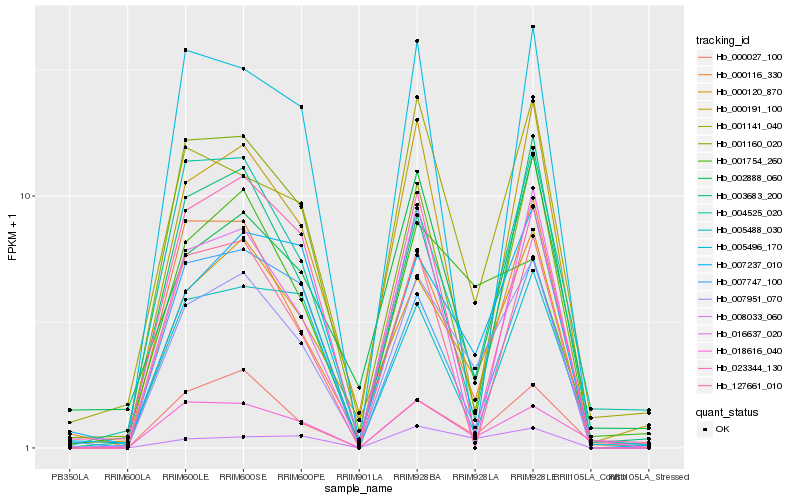
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007951_070 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 2 |
Hb_000120_870 |
0.1029695268 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 3 |
Hb_007237_010 |
0.1425484241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000116_330 |
0.1469099023 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 5 |
Hb_001141_040 |
0.1547547449 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 6 |
Hb_008033_060 |
0.1582769219 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 7 |
Hb_018616_040 |
0.1599060479 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000027_100 |
0.1611436152 |
- |
- |
hypothetical protein VITISV_039936 [Vitis vinifera] |
| 9 |
Hb_001754_260 |
0.161347434 |
- |
- |
PREDICTED: uncharacterized protein LOC105643409 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000191_100 |
0.1629641589 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_003683_200 |
0.1739990852 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 12 |
Hb_004525_020 |
0.1746488057 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |
| 13 |
Hb_007747_100 |
0.1809346893 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 14 |
Hb_127661_010 |
0.1854389607 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_016637_020 |
0.1859659718 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 16 |
Hb_001160_020 |
0.1861148844 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 17 |
Hb_002888_060 |
0.187003851 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_023344_130 |
0.1870694613 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_005488_030 |
0.189946207 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 20 |
Hb_005496_170 |
0.191740185 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |