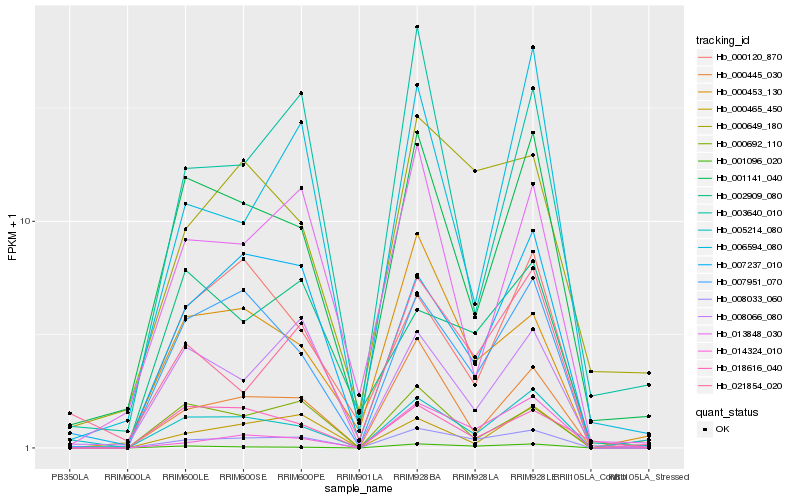
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008033_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 2 |
Hb_007951_070 |
0.1582769219 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 3 |
Hb_001096_020 |
0.159409087 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Populus euphratica] |
| 4 |
Hb_000649_180 |
0.1769944272 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 5 |
Hb_007237_010 |
0.1841346488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000445_030 |
0.1843713937 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 7 |
Hb_000692_110 |
0.1863958422 |
- |
- |
PREDICTED: NEP1-interacting protein-like 1 [Jatropha curcas] |
| 8 |
Hb_001141_040 |
0.1894999747 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 9 |
Hb_000453_130 |
0.1909157091 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_021854_020 |
0.1933770031 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 11 |
Hb_008066_080 |
0.1992115903 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 12 |
Hb_000465_450 |
0.2050533802 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 13 |
Hb_013848_030 |
0.2145324478 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 14 |
Hb_005214_080 |
0.2154709029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_014324_010 |
0.21706621 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 16 |
Hb_002909_080 |
0.2173368706 |
- |
- |
inositol or phosphatidylinositol kinase, putative [Ricinus communis] |
| 17 |
Hb_000120_870 |
0.2218106351 |
rubber biosynthesis |
Gene Name: CPT6 |
- |
| 18 |
Hb_003640_010 |
0.2238845802 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 19 |
Hb_018616_040 |
0.2262074621 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_006594_080 |
0.2268083109 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |