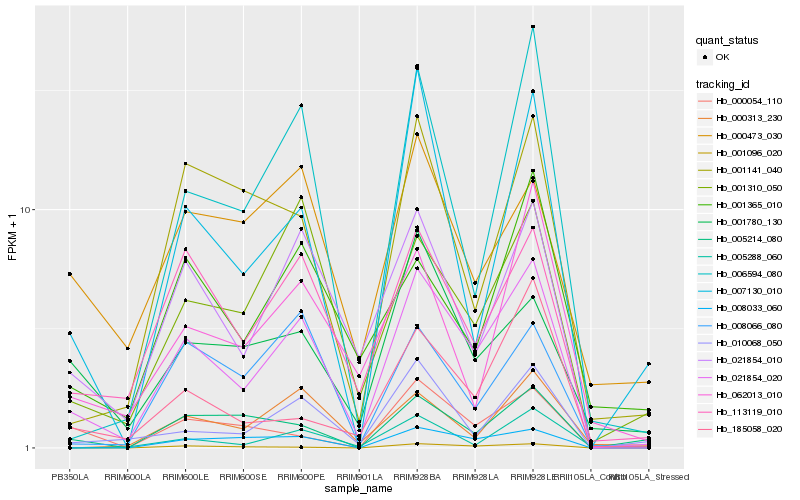
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021854_020 |
0.0 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 2 |
Hb_021854_010 |
0.1535460659 |
- |
- |
- |
| 3 |
Hb_113119_010 |
0.1545620953 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_001310_050 |
0.1828584683 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 5 |
Hb_010068_050 |
0.1896785553 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 6 |
Hb_006594_080 |
0.1910194184 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 7 |
Hb_008033_060 |
0.1933770031 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 8 |
Hb_007130_010 |
0.199442052 |
- |
- |
- |
| 9 |
Hb_000313_230 |
0.2036620727 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 10 |
Hb_008066_080 |
0.2064295251 |
- |
- |
PREDICTED: GDSL esterase/lipase 1-like [Jatropha curcas] |
| 11 |
Hb_005214_080 |
0.2077690106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001096_020 |
0.2083402561 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Populus euphratica] |
| 13 |
Hb_001141_040 |
0.2088609707 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 14 |
Hb_001780_130 |
0.2103754293 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 15 |
Hb_001365_010 |
0.212150272 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 16 |
Hb_185058_020 |
0.2122529268 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 17 |
Hb_005288_060 |
0.2153264634 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 18 |
Hb_000054_110 |
0.2159513604 |
- |
- |
PREDICTED: uncharacterized protein LOC105350810 [Fragaria vesca subsp. vesca] |
| 19 |
Hb_062013_010 |
0.2203832227 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_000473_030 |
0.2208412508 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |