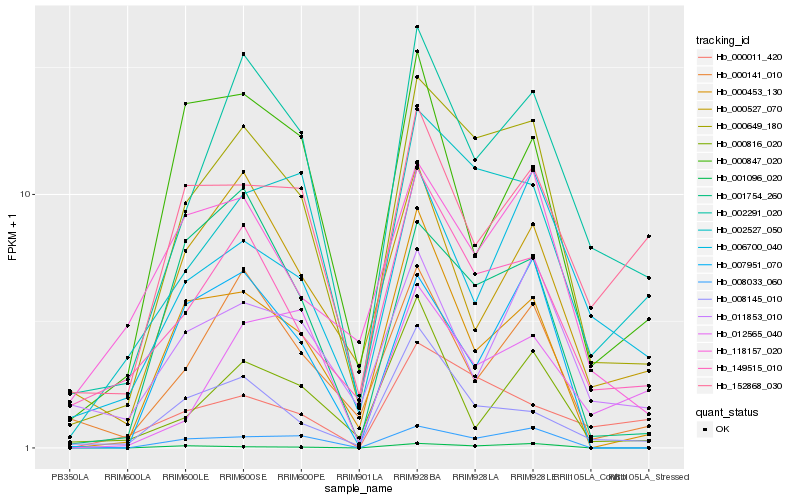
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_180 |
0.0 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 2 |
Hb_002291_020 |
0.1458579496 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_002527_050 |
0.155735656 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 4 |
Hb_149515_010 |
0.1568467677 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 5 |
Hb_008033_060 |
0.1769944272 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 6 |
Hb_000453_130 |
0.1853259948 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_001754_260 |
0.1892472764 |
- |
- |
PREDICTED: uncharacterized protein LOC105643409 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000141_010 |
0.1919420653 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 9 |
Hb_008145_010 |
0.1944602613 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_118157_020 |
0.1947511795 |
- |
- |
hypothetical protein MTR_3g456110 [Medicago truncatula] |
| 11 |
Hb_006700_040 |
0.1974599736 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 12 |
Hb_000011_420 |
0.2094203919 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29660-like [Citrus sinensis] |
| 13 |
Hb_000527_070 |
0.2099172983 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 14 |
Hb_152868_030 |
0.216505309 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 15 |
Hb_011853_010 |
0.2198371168 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 16 |
Hb_007951_070 |
0.2200844208 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 17 |
Hb_000847_020 |
0.2210896707 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 18 |
Hb_001096_020 |
0.2257190505 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000816_020 |
0.232180211 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_012565_040 |
0.2326230339 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |