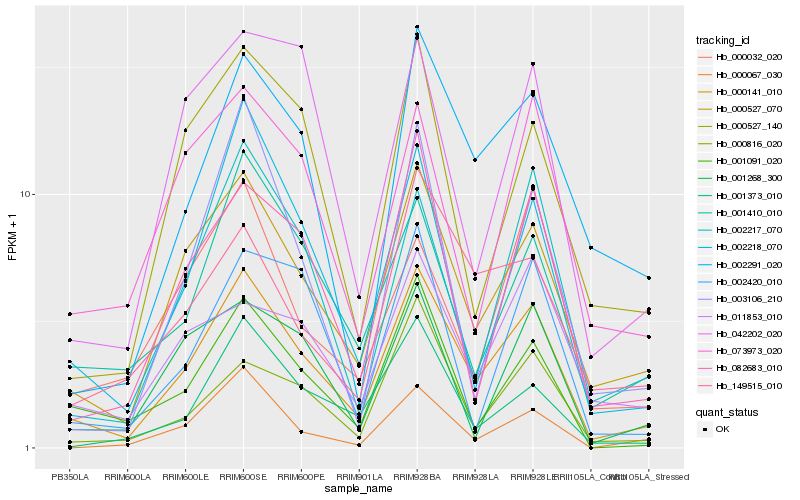
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000141_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 2 |
Hb_000527_070 |
0.0931865632 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 3 |
Hb_002218_070 |
0.1488325693 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 4 |
Hb_001410_010 |
0.1529444991 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 5 |
Hb_082683_010 |
0.1541905166 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_002291_020 |
0.1564001913 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_001091_020 |
0.1566192483 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 8 |
Hb_002217_070 |
0.1570675233 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 9 |
Hb_000527_140 |
0.1645023315 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 10 |
Hb_000816_020 |
0.1662858283 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_001373_010 |
0.1673055056 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_073973_020 |
0.1707473357 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 13 |
Hb_149515_010 |
0.1709047043 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 14 |
Hb_011853_010 |
0.1710418256 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 15 |
Hb_001268_300 |
0.1762924063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000067_030 |
0.1780851559 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 17 |
Hb_003106_210 |
0.1804665816 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74B1-like [Jatropha curcas] |
| 18 |
Hb_042202_020 |
0.1805558209 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 19 |
Hb_002420_010 |
0.1808945558 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 20 |
Hb_000032_020 |
0.1835682028 |
- |
- |
Centromeric protein E, putative [Ricinus communis] |