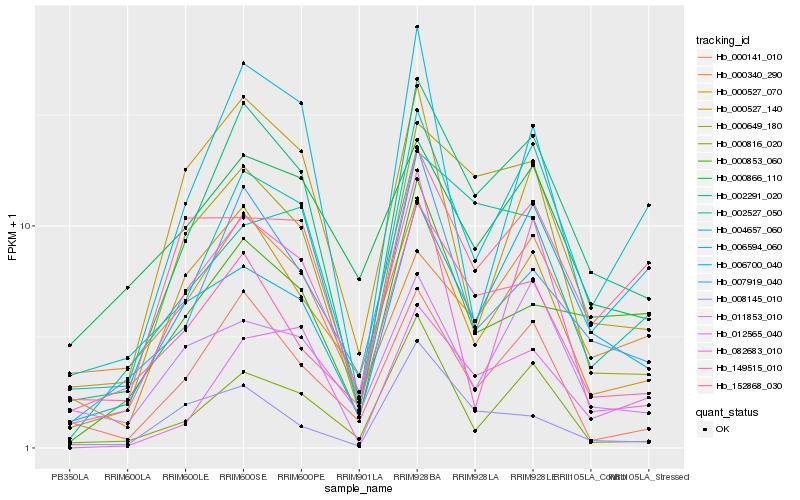
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002291_020 |
0.0 |
- |
- |
PREDICTED: glutamate synthase 1 [NADH], chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000649_180 |
0.1458579496 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 3 |
Hb_007919_040 |
0.1549033074 |
- |
- |
PREDICTED: protein LYK5 [Jatropha curcas] |
| 4 |
Hb_000141_010 |
0.1564001913 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 5 |
Hb_149515_010 |
0.1579552009 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 6 |
Hb_012565_040 |
0.1596289563 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 7 |
Hb_000527_070 |
0.1651567577 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 8 |
Hb_006700_040 |
0.1708780419 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 9 |
Hb_002527_050 |
0.1757278803 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 10 |
Hb_004657_060 |
0.1812629155 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_000853_060 |
0.1833217973 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_006594_060 |
0.1855273633 |
transcription factor |
TF Family: HSF |
PREDICTED: heat shock factor protein HSF24 [Jatropha curcas] |
| 13 |
Hb_000816_020 |
0.1857415662 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_152868_030 |
0.1885687517 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 15 |
Hb_000527_140 |
0.1906419073 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 16 |
Hb_011853_010 |
0.1957970788 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 17 |
Hb_082683_010 |
0.2017228778 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_000866_110 |
0.2017988112 |
- |
- |
PREDICTED: uncharacterized protein LOC105642980 isoform X2 [Jatropha curcas] |
| 19 |
Hb_008145_010 |
0.2024232949 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 20 |
Hb_000340_290 |
0.203207682 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |