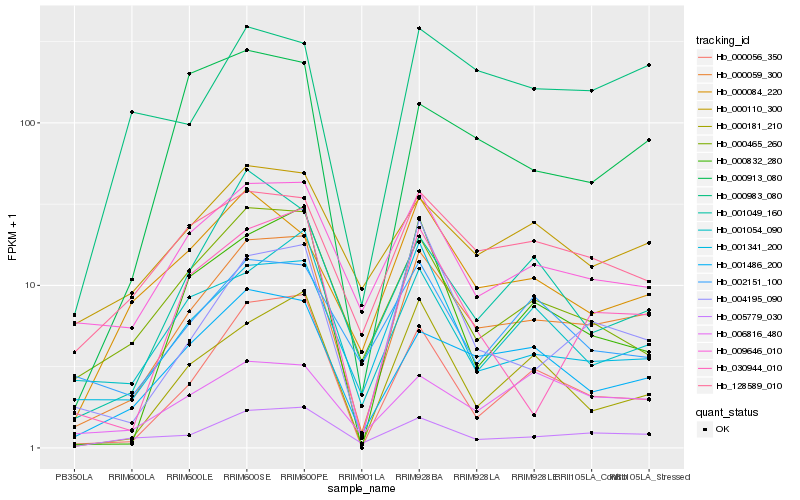
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_300 |
0.0 |
- |
- |
caspase, putative [Ricinus communis] |
| 2 |
Hb_001486_200 |
0.1261342315 |
- |
- |
PREDICTED: uncharacterized protein LOC105632606 [Jatropha curcas] |
| 3 |
Hb_005779_030 |
0.1279232565 |
- |
- |
PREDICTED: nuclear pore complex protein NUP62 [Jatropha curcas] |
| 4 |
Hb_000056_350 |
0.1387875087 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 5 |
Hb_009646_010 |
0.148871527 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 6 |
Hb_001341_200 |
0.1515725599 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 7 |
Hb_000181_210 |
0.1572304991 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 8 |
Hb_128589_010 |
0.1586794768 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_000465_260 |
0.1589571204 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 10 |
Hb_000110_300 |
0.162262727 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |
| 11 |
Hb_004195_090 |
0.1638286493 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 12 |
Hb_001054_090 |
0.1654579551 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 13 |
Hb_030944_010 |
0.1702825518 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 14 |
Hb_000832_280 |
0.170813314 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 15 |
Hb_000913_080 |
0.171571148 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 11 [Jatropha curcas] |
| 16 |
Hb_006816_480 |
0.1737355604 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001049_160 |
0.174610918 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 18 |
Hb_000084_220 |
0.1759637267 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 19 |
Hb_000983_080 |
0.1771414324 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 20 |
Hb_002151_100 |
0.1785960275 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |