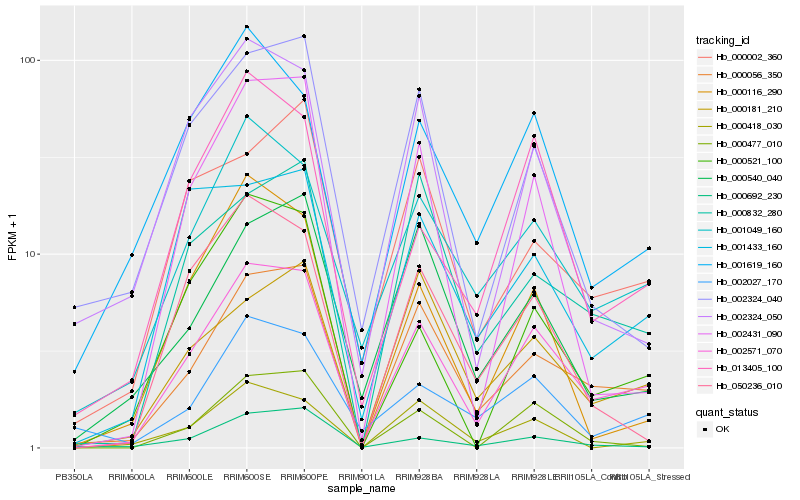
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002571_070 |
0.0 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 2 |
Hb_000056_350 |
0.1071823304 |
- |
- |
PREDICTED: uncharacterized protein LOC105630437 [Jatropha curcas] |
| 3 |
Hb_001049_160 |
0.1417532956 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 4 |
Hb_013405_100 |
0.160224329 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 5 |
Hb_000477_010 |
0.1643100409 |
- |
- |
PREDICTED: potassium channel AKT1-like [Jatropha curcas] |
| 6 |
Hb_000521_100 |
0.1691832093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002431_090 |
0.1708910867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000692_230 |
0.1712801872 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 9 |
Hb_000418_030 |
0.1744815896 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 10 |
Hb_001619_160 |
0.1761456876 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 11 |
Hb_002027_170 |
0.1764460759 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 12 |
Hb_050236_010 |
0.1767109472 |
- |
- |
hypothetical protein EUGRSUZ_E02960, partial [Eucalyptus grandis] |
| 13 |
Hb_000181_210 |
0.1796016336 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_001433_160 |
0.1804277373 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 15 |
Hb_002324_050 |
0.1806195122 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 16 |
Hb_000832_280 |
0.1807911591 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 17 |
Hb_002324_040 |
0.1819814601 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 18 |
Hb_000116_290 |
0.1821365919 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 19 |
Hb_000002_360 |
0.182990318 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 20 |
Hb_000540_040 |
0.1838135139 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |