| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
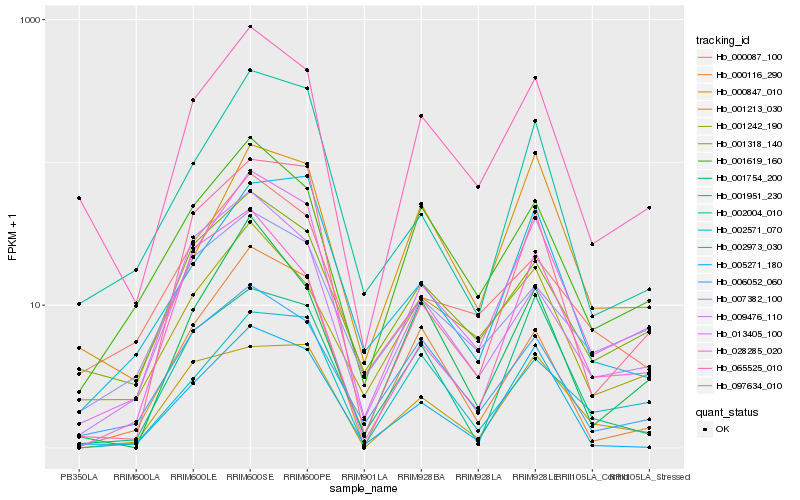
Hb_013405_100 |
0.0 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 2 |
Hb_065525_010 |
0.1146060648 |
- |
- |
hypothetical protein JCGZ_01481 [Jatropha curcas] |
| 3 |
Hb_002004_010 |
0.1229287283 |
- |
- |
CBL-interacting protein kinase 9 [Populus trichocarpa] |
| 4 |
Hb_097634_010 |
0.1384901365 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 5 |
Hb_001318_140 |
0.1390875384 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 6 |
Hb_001619_160 |
0.1397776357 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 7 |
Hb_005271_180 |
0.1470433411 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 8 |
Hb_002973_030 |
0.1495841753 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000087_100 |
0.1524076811 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 10 |
Hb_006052_060 |
0.1543329511 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 11 |
Hb_001754_200 |
0.1550946804 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 12 |
Hb_007382_100 |
0.1595394998 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000116_290 |
0.1595577091 |
- |
- |
PREDICTED: proton-coupled amino acid transporter 3 [Jatropha curcas] |
| 14 |
Hb_002571_070 |
0.160224329 |
- |
- |
PREDICTED: cation/calcium exchanger 2 [Jatropha curcas] |
| 15 |
Hb_001242_190 |
0.1611868247 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 16 |
Hb_009476_110 |
0.1614680928 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 17 |
Hb_000847_010 |
0.1625510263 |
- |
- |
PREDICTED: uncharacterized protein LOC102608064 [Citrus sinensis] |
| 18 |
Hb_028285_020 |
0.1640594948 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_001213_030 |
0.1641663282 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 20 |
Hb_001951_230 |
0.1651043279 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |