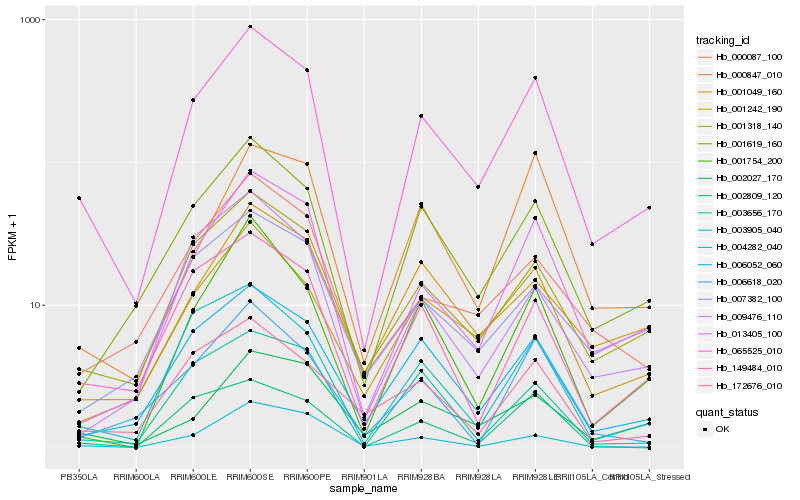
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065525_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01481 [Jatropha curcas] |
| 2 |
Hb_001318_140 |
0.1136397408 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 3 |
Hb_001242_190 |
0.1141491614 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 4 |
Hb_013405_100 |
0.1146060648 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 5 |
Hb_001619_160 |
0.1234821673 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 6 |
Hb_006052_060 |
0.1254970839 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 7 |
Hb_149484_010 |
0.1436673716 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 8 |
Hb_003905_040 |
0.1442931189 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 9 |
Hb_001754_200 |
0.1482741957 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 10 |
Hb_000087_100 |
0.1521719546 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 11 |
Hb_002027_170 |
0.1611018 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RKF3 [Jatropha curcas] |
| 12 |
Hb_172676_010 |
0.166091596 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 13 |
Hb_007382_100 |
0.1664519227 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_003656_170 |
0.1667890481 |
- |
- |
PREDICTED: phenylalanine ammonia-lyase-like [Jatropha curcas] |
| 15 |
Hb_000847_010 |
0.168060301 |
- |
- |
PREDICTED: uncharacterized protein LOC102608064 [Citrus sinensis] |
| 16 |
Hb_002809_120 |
0.16875357 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 17 |
Hb_006618_020 |
0.1688143645 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 18 |
Hb_009476_110 |
0.1717492233 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 19 |
Hb_001049_160 |
0.1718194939 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 20 |
Hb_004282_040 |
0.1726217668 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |