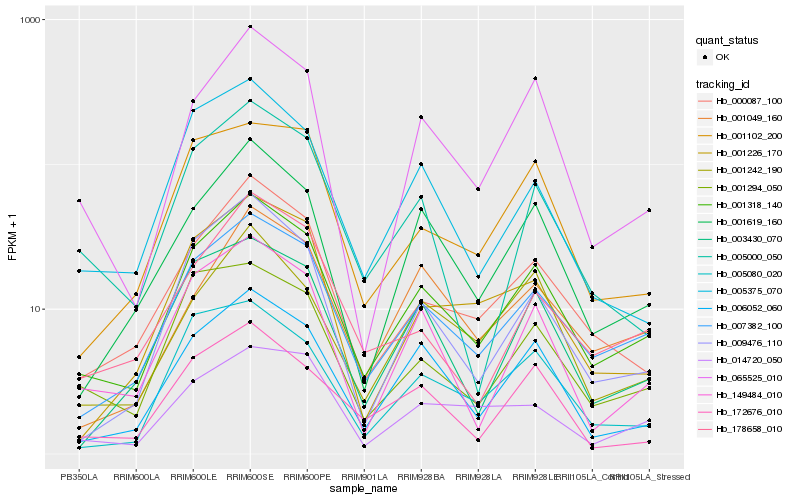
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_140 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 2 |
Hb_007382_100 |
0.0656723794 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000087_100 |
0.1024653466 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 4 |
Hb_006052_060 |
0.104186822 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 5 |
Hb_001619_160 |
0.1104586987 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 6 |
Hb_065525_010 |
0.1136397408 |
- |
- |
hypothetical protein JCGZ_01481 [Jatropha curcas] |
| 7 |
Hb_149484_010 |
0.1191423795 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor-like [Eucalyptus grandis] |
| 8 |
Hb_009476_110 |
0.1225110069 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 9 |
Hb_001242_190 |
0.1249980312 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 1 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001294_050 |
0.1289775758 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 11 |
Hb_003430_070 |
0.1315757203 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 12 |
Hb_005080_020 |
0.1335516147 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 13 |
Hb_178658_010 |
0.1351539047 |
- |
- |
PREDICTED: uncharacterized protein LOC105641396 [Jatropha curcas] |
| 14 |
Hb_172676_010 |
0.1356898325 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 15 |
Hb_001049_160 |
0.1362751408 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 16 |
Hb_014720_050 |
0.1366653126 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 17 |
Hb_001226_170 |
0.1369895591 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 18 |
Hb_001102_200 |
0.1369905249 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 19 |
Hb_005375_070 |
0.1374329266 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein LHY [Jatropha curcas] |
| 20 |
Hb_005000_050 |
0.1383415698 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |