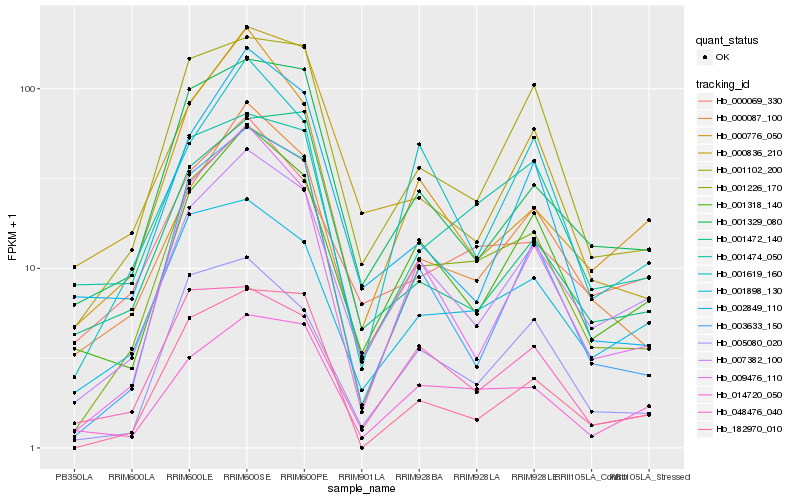
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001226_170 |
0.0 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 2 |
Hb_000087_100 |
0.1111839449 |
- |
- |
PREDICTED: cellulose synthase-like protein D3 [Jatropha curcas] |
| 3 |
Hb_003633_150 |
0.1232994224 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 4 |
Hb_009476_110 |
0.1262331234 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 5 |
Hb_001102_200 |
0.1285245485 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 6 |
Hb_007382_100 |
0.1299790691 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_182970_010 |
0.1315670701 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 8 |
Hb_001318_140 |
0.1369895591 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 9 |
Hb_014720_050 |
0.1370077137 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 10 |
Hb_001329_080 |
0.1389493011 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 11 |
Hb_005080_020 |
0.1407277898 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 12 |
Hb_000776_050 |
0.1580549525 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 13 |
Hb_001474_050 |
0.1581528257 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001898_130 |
0.1589811449 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000836_210 |
0.1605269183 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 16 |
Hb_001619_160 |
0.1616053675 |
transcription factor |
TF Family: ERF |
DREB1p [Hevea brasiliensis] |
| 17 |
Hb_048476_040 |
0.1632150917 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 18 |
Hb_002849_110 |
0.1632250244 |
- |
- |
hypothetical protein JCGZ_14362 [Jatropha curcas] |
| 19 |
Hb_001472_140 |
0.1641634456 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 20 |
Hb_000069_330 |
0.1659799596 |
- |
- |
protein with unknown function [Ricinus communis] |