| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
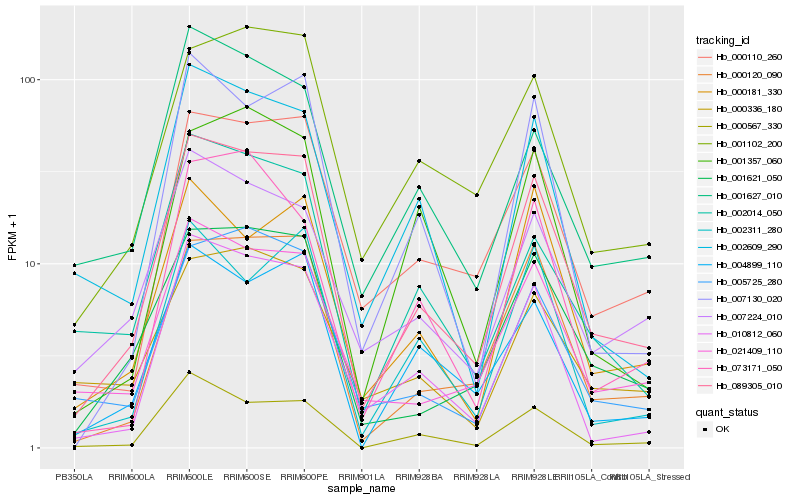
Hb_089305_010 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 2 |
Hb_000120_090 |
0.095938011 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 3 |
Hb_001621_050 |
0.1055976343 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 4 |
Hb_000567_330 |
0.1088926482 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 5 |
Hb_010812_060 |
0.1102315108 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 6 |
Hb_000110_260 |
0.110965745 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 7 |
Hb_021409_110 |
0.1143383544 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_005725_280 |
0.118227839 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 9 |
Hb_001102_200 |
0.1256274056 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 10 |
Hb_004899_110 |
0.1277279014 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 11 |
Hb_007130_020 |
0.1282704873 |
- |
- |
- |
| 12 |
Hb_007224_010 |
0.1319448866 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 13 |
Hb_002609_290 |
0.1355229248 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 14 |
Hb_001627_010 |
0.1401439138 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 15 |
Hb_000181_330 |
0.1418820325 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 16 |
Hb_002014_050 |
0.1434181182 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 17 |
Hb_002311_280 |
0.1455053167 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 18 |
Hb_001357_060 |
0.1458474799 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 19 |
Hb_073171_050 |
0.146437971 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 20 |
Hb_000336_180 |
0.1478841497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |