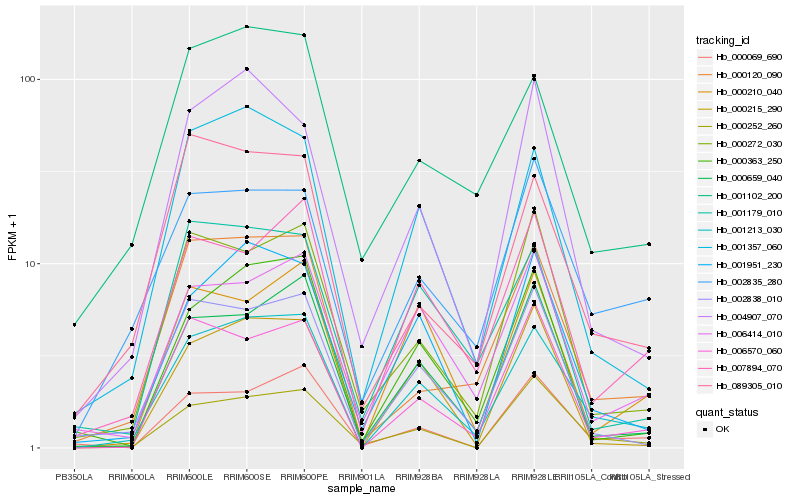
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001213_030 |
0.0 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 2 |
Hb_001357_060 |
0.1239731275 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 3 |
Hb_001951_230 |
0.1328962101 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 4 |
Hb_000659_040 |
0.1380069928 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_002838_010 |
0.1417538218 |
- |
- |
AGPase large subunit protein [Hevea brasiliensis] |
| 6 |
Hb_000363_250 |
0.1419749404 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 7 |
Hb_000120_090 |
0.142277806 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 8 |
Hb_007894_070 |
0.1465234592 |
- |
- |
PREDICTED: uncharacterized protein LOC105632955 [Jatropha curcas] |
| 9 |
Hb_000252_260 |
0.1469100129 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 10 |
Hb_006570_060 |
0.1513797477 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 11 |
Hb_001102_200 |
0.154819278 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 12 |
Hb_006414_010 |
0.1548240602 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 13 |
Hb_000215_290 |
0.1576202719 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002835_280 |
0.157866377 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 15 |
Hb_000210_040 |
0.1587776437 |
- |
- |
PREDICTED: putative HVA22-like protein g [Jatropha curcas] |
| 16 |
Hb_089305_010 |
0.1589297873 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 17 |
Hb_000272_030 |
0.1595798966 |
- |
- |
PREDICTED: WD repeat-containing protein 76 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000069_690 |
0.1596131365 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF6 [Jatropha curcas] |
| 19 |
Hb_001179_010 |
0.1624899655 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 20 |
Hb_004907_070 |
0.1626643114 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |