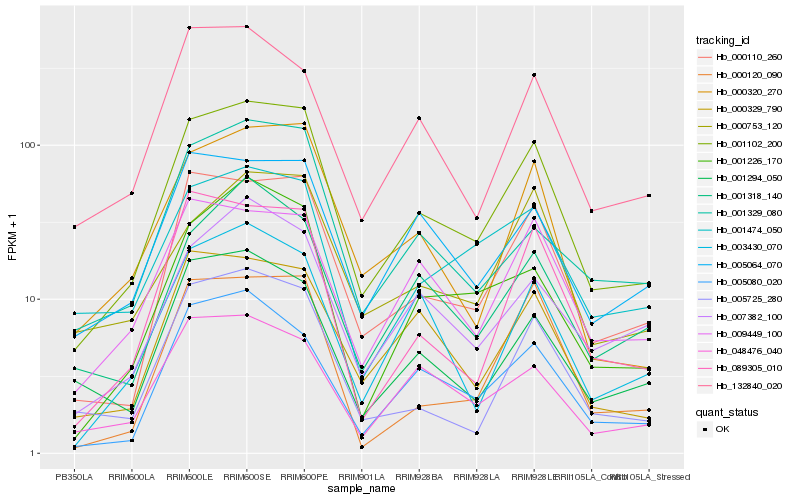
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_200 |
0.0 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 2 |
Hb_000110_260 |
0.0902981818 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 3 |
Hb_000320_270 |
0.1079957324 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000329_790 |
0.1207747048 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 5 |
Hb_005064_070 |
0.1238588175 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 6 |
Hb_005725_280 |
0.1252007566 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 7 |
Hb_089305_010 |
0.1256274056 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 8 |
Hb_009449_100 |
0.1262697255 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 9 |
Hb_005080_020 |
0.1279080584 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 10 |
Hb_001226_170 |
0.1285245485 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 11 |
Hb_001474_050 |
0.1319593634 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000120_090 |
0.1327629072 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 13 |
Hb_003430_070 |
0.1328664387 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 14 |
Hb_000753_120 |
0.133725147 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 15 |
Hb_001294_050 |
0.1342789985 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 16 |
Hb_001329_080 |
0.1346408527 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 17 |
Hb_132840_020 |
0.1359323619 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 18 |
Hb_007382_100 |
0.1364044703 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 19 |
Hb_048476_040 |
0.1368692122 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 20 |
Hb_001318_140 |
0.1369905249 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |