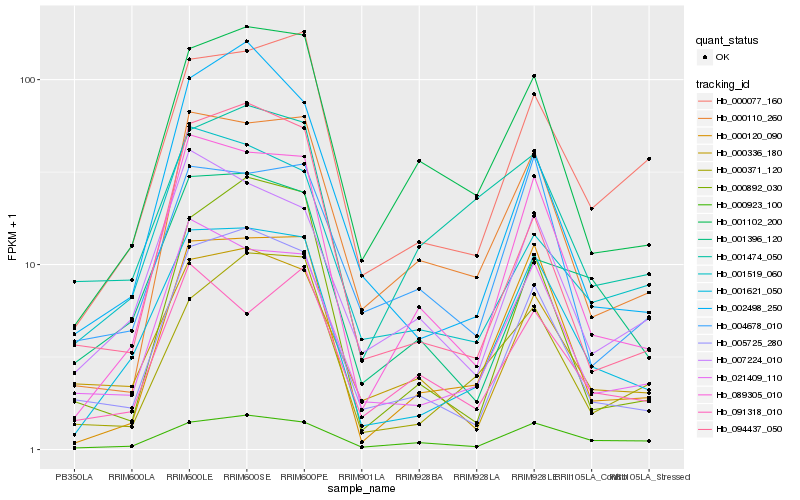
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 2 |
Hb_089305_010 |
0.1055976343 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 3 |
Hb_000120_090 |
0.1194932372 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 4 |
Hb_021409_110 |
0.1211641881 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 5 |
Hb_005725_280 |
0.1327914791 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 6 |
Hb_000077_160 |
0.1394722995 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 7 |
Hb_001102_200 |
0.1445357865 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 8 |
Hb_000336_180 |
0.1504350039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000110_260 |
0.1534996665 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 10 |
Hb_007224_010 |
0.1555059447 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 11 |
Hb_000923_100 |
0.1557563985 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 12 |
Hb_001396_120 |
0.1607505775 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 13 |
Hb_000371_120 |
0.1631515732 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_001519_060 |
0.1679578301 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_004678_010 |
0.1724250078 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 16 |
Hb_002498_250 |
0.1796943197 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_091318_010 |
0.1813263813 |
- |
- |
hypothetical protein POPTR_0001s09520g [Populus trichocarpa] |
| 18 |
Hb_001474_050 |
0.1835239882 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 19 |
Hb_094437_050 |
0.1837265614 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000892_030 |
0.1839529943 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |