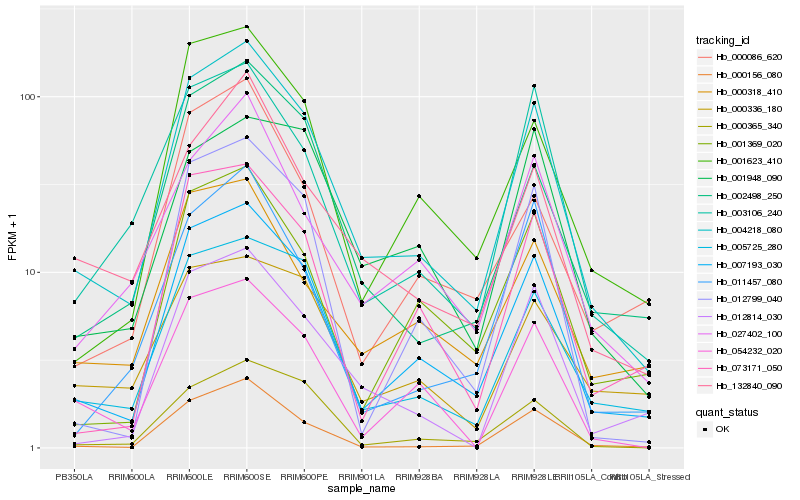
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |
| 2 |
Hb_007193_030 |
0.0673319883 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005725_280 |
0.1137800621 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 4 |
Hb_002498_250 |
0.1184845235 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_000156_080 |
0.1257677463 |
- |
- |
Potassium channel KAT2, putative [Ricinus communis] |
| 6 |
Hb_000365_340 |
0.1264971529 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 7 |
Hb_003106_240 |
0.1273093876 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 8 |
Hb_001623_410 |
0.1307248288 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_132840_090 |
0.1315792364 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_012814_030 |
0.1355723142 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 11 |
Hb_054232_020 |
0.1372595957 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 12 |
Hb_027402_100 |
0.1373765003 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 13 |
Hb_012799_040 |
0.1443937989 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 14 |
Hb_000318_410 |
0.1453000751 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_001369_020 |
0.1464253585 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 16 |
Hb_073171_050 |
0.1479564832 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 17 |
Hb_011457_080 |
0.1479959982 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000086_620 |
0.1588983772 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001948_090 |
0.159473967 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0005s14540g [Populus trichocarpa] |
| 20 |
Hb_000336_180 |
0.1597864914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |