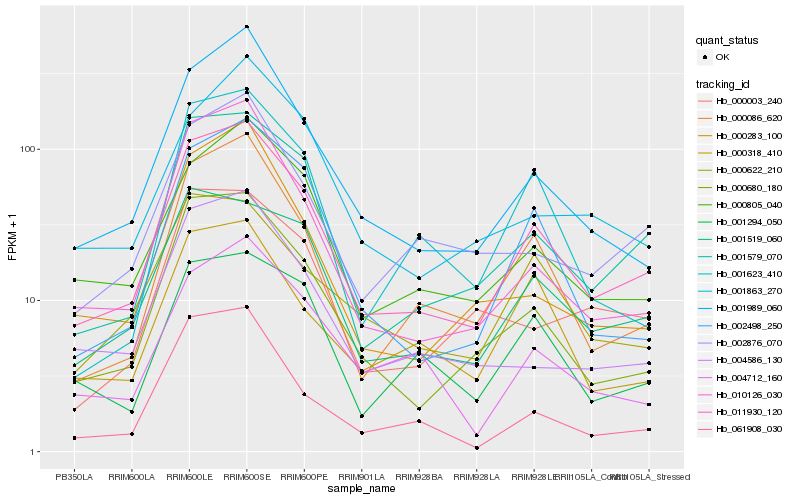
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011930_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001579_070 |
0.0964474715 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 3 |
Hb_000680_180 |
0.1001363324 |
- |
- |
hypothetical protein JCGZ_15248 [Jatropha curcas] |
| 4 |
Hb_000086_620 |
0.1044379858 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001989_060 |
0.1132130132 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105634562 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000805_040 |
0.1165844498 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 7 |
Hb_002498_250 |
0.1197954011 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001519_060 |
0.1200312905 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_010126_030 |
0.1273970103 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002876_070 |
0.1300845208 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 11 |
Hb_004586_130 |
0.134026915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004712_160 |
0.1350933843 |
- |
- |
hypothetical protein CICLE_v10024612mg [Citrus clementina] |
| 13 |
Hb_001294_050 |
0.1366167895 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 14 |
Hb_061908_030 |
0.1382100978 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 15 |
Hb_001623_410 |
0.1387536129 |
- |
- |
PREDICTED: beta-amylase 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000283_100 |
0.1393830121 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000622_210 |
0.1411039755 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 18 |
Hb_000318_410 |
0.1427078662 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 19 |
Hb_001863_270 |
0.1475927477 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 20 |
Hb_000003_240 |
0.1479311667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |