| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
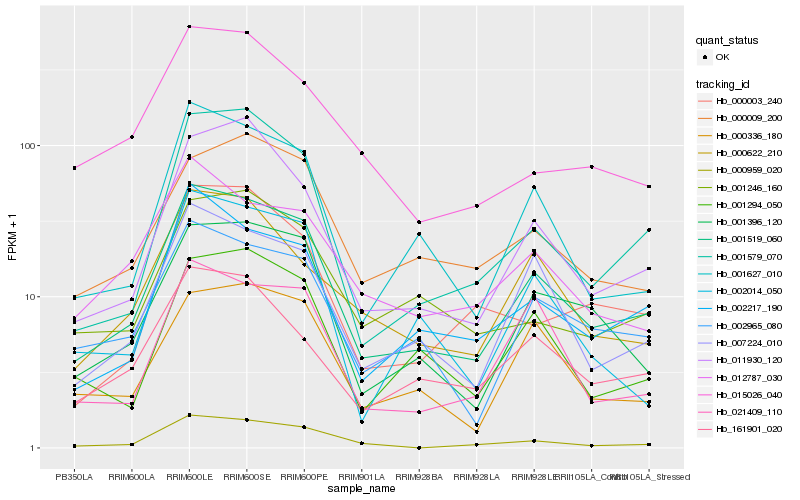
Hb_001519_060 |
0.0 |
- |
- |
PREDICTED: glutamyl-tRNA reductase 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_007224_010 |
0.1090730303 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 3 |
Hb_001627_010 |
0.1093788896 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 4 |
Hb_002965_080 |
0.1135767 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 5 |
Hb_001396_120 |
0.1138240455 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 6 |
Hb_001579_070 |
0.1153388255 |
- |
- |
PREDICTED: uncharacterized protein LOC105644596 [Jatropha curcas] |
| 7 |
Hb_002217_190 |
0.1158708327 |
- |
- |
invertase [Hevea brasiliensis] |
| 8 |
Hb_011930_120 |
0.1200312905 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000959_020 |
0.1306206554 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_161901_020 |
0.1328004295 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Vitis vinifera] |
| 11 |
Hb_000336_180 |
0.1344835935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001294_050 |
0.1346935027 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 13 |
Hb_000622_210 |
0.1386696209 |
transcription factor |
TF Family: bZIP |
PREDICTED: common plant regulatory factor 1-like isoform X3 [Jatropha curcas] |
| 14 |
Hb_021409_110 |
0.1398737971 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_000009_200 |
0.1410451088 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 16 |
Hb_002014_050 |
0.1419518899 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 17 |
Hb_001246_160 |
0.1432616075 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 18 |
Hb_012787_030 |
0.1435187268 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 19 |
Hb_015026_040 |
0.1474255641 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 28 homolog 2 [Jatropha curcas] |
| 20 |
Hb_000003_240 |
0.1475237435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |