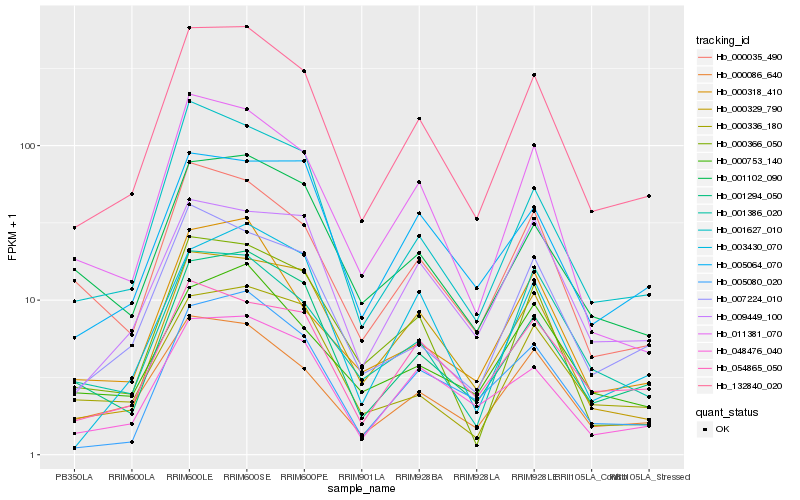
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132840_020 |
0.0 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 2 |
Hb_001294_050 |
0.0896403222 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 3 |
Hb_000086_640 |
0.0908853842 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_048476_040 |
0.0983687779 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 5 |
Hb_011381_070 |
0.0986367846 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 6 |
Hb_001627_010 |
0.1009724938 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 7 |
Hb_000329_790 |
0.1031688114 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 8 |
Hb_007224_010 |
0.10789956 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 9 |
Hb_005080_020 |
0.1083677772 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 10 |
Hb_000035_490 |
0.108627819 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 11 |
Hb_009449_100 |
0.1124891228 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 12 |
Hb_054865_050 |
0.1133568539 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000318_410 |
0.1144966465 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 14 |
Hb_001386_020 |
0.1151822511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000753_140 |
0.1157706113 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 16 |
Hb_001102_090 |
0.1157721605 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 17 |
Hb_000366_050 |
0.1173335123 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 18 |
Hb_005064_070 |
0.1180903954 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 19 |
Hb_000336_180 |
0.118763482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003430_070 |
0.119905191 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |