| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
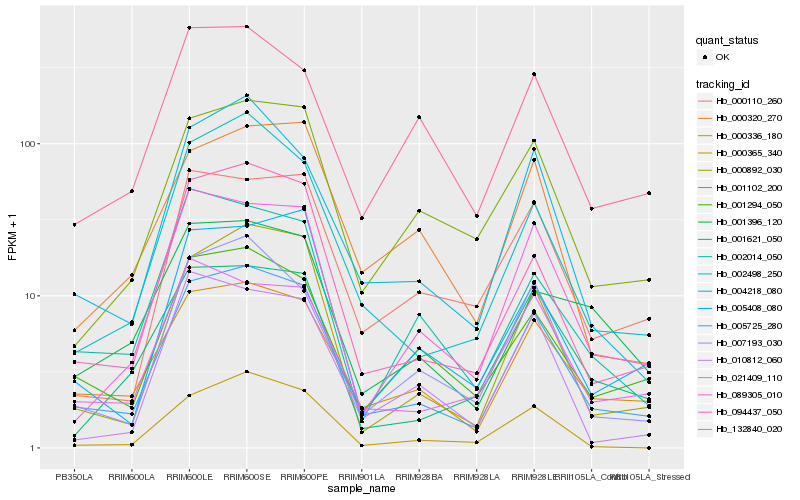
Hb_005725_280 |
0.0 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 2 |
Hb_000336_180 |
0.0908864563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_094437_050 |
0.1011649739 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000892_030 |
0.1065887438 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 5 |
Hb_002498_250 |
0.1110263956 |
- |
- |
5'-adenylylsulfate reductase 1, chloroplast precursor, putative [Ricinus communis] |
| 6 |
Hb_004218_080 |
0.1137800621 |
- |
- |
PREDICTED: uncharacterized protein LOC105631207 [Jatropha curcas] |
| 7 |
Hb_007193_030 |
0.1140686126 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ706 isoform X1 [Jatropha curcas] |
| 8 |
Hb_089305_010 |
0.118227839 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 9 |
Hb_001294_050 |
0.1193085104 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 10 |
Hb_001102_200 |
0.1252007566 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 11 |
Hb_000365_340 |
0.1294313296 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 12 |
Hb_021409_110 |
0.131105171 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_001621_050 |
0.1327914791 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 14 |
Hb_005408_080 |
0.133758205 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 15 |
Hb_000110_260 |
0.1386283526 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 16 |
Hb_000320_270 |
0.1390429416 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002014_050 |
0.1407257562 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 isoform X2 [Populus euphratica] |
| 18 |
Hb_132840_020 |
0.1457696665 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 19 |
Hb_001396_120 |
0.1459015722 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 20 |
Hb_010812_060 |
0.1463422075 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |