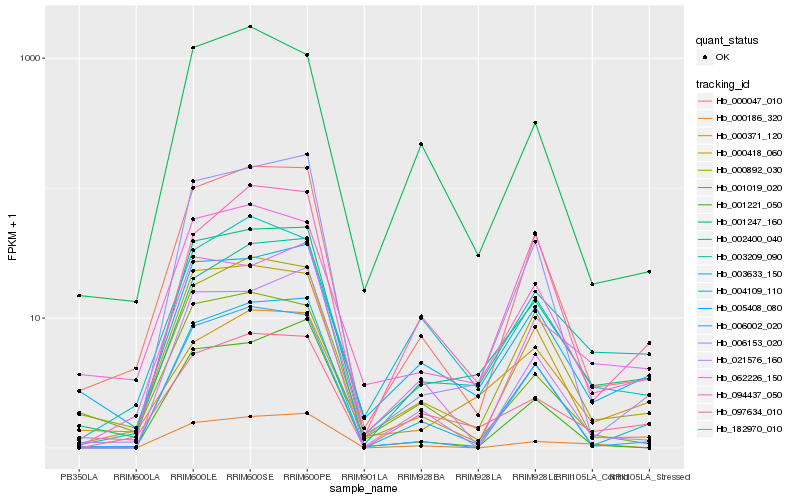
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002400_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637633 [Jatropha curcas] |
| 2 |
Hb_005408_080 |
0.1044158751 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 3 |
Hb_000047_010 |
0.1074663162 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 4 |
Hb_000892_030 |
0.1102427731 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 5 |
Hb_094437_050 |
0.1127366458 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 6 |
Hb_182970_010 |
0.1149391463 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 7 |
Hb_004109_110 |
0.1201131227 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 8 |
Hb_000418_060 |
0.1284850027 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 9 |
Hb_006153_020 |
0.1326540493 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 10 |
Hb_003209_090 |
0.1352685729 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 11 |
Hb_062226_150 |
0.1442397065 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003633_150 |
0.1444650951 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 13 |
Hb_006002_020 |
0.145872892 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 14 |
Hb_001247_160 |
0.1479784072 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 15 |
Hb_001019_020 |
0.1484814713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_021576_160 |
0.1495991018 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_097634_010 |
0.1553320004 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 18 |
Hb_000371_120 |
0.1594186448 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000186_320 |
0.1599760344 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_001221_050 |
0.1607395819 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At3g03770 [Jatropha curcas] |