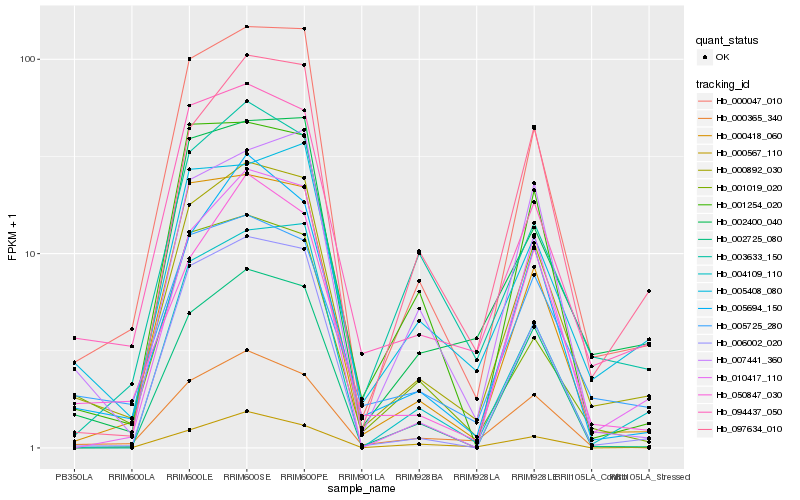
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000892_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 2 |
Hb_000047_010 |
0.073691361 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 3 |
Hb_094437_050 |
0.0990167439 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005725_280 |
0.1065887438 |
- |
- |
PREDICTED: sodium-coupled neutral amino acid transporter 5-like [Jatropha curcas] |
| 5 |
Hb_005694_150 |
0.107029244 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 6 |
Hb_000365_340 |
0.1072483244 |
- |
- |
PREDICTED: uncharacterized protein LOC105649067 [Jatropha curcas] |
| 7 |
Hb_002400_040 |
0.1102427731 |
- |
- |
PREDICTED: uncharacterized protein LOC105637633 [Jatropha curcas] |
| 8 |
Hb_050847_030 |
0.1142025802 |
- |
- |
PREDICTED: cytochrome P450 714A1-like [Populus euphratica] |
| 9 |
Hb_097634_010 |
0.1158570914 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 10 |
Hb_000418_060 |
0.1164745937 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 11 |
Hb_005408_080 |
0.1219118202 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 12 |
Hb_006002_020 |
0.1230437896 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 13 |
Hb_010417_110 |
0.124148844 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002725_080 |
0.1242184104 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_004109_110 |
0.1284248536 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |
| 16 |
Hb_001019_020 |
0.1301107475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007441_360 |
0.1328452781 |
- |
- |
PREDICTED: uncharacterized protein LOC105643270 [Jatropha curcas] |
| 18 |
Hb_000567_110 |
0.1328601245 |
- |
- |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 19 |
Hb_003633_150 |
0.1334632198 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 20 |
Hb_001254_020 |
0.1357427887 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |