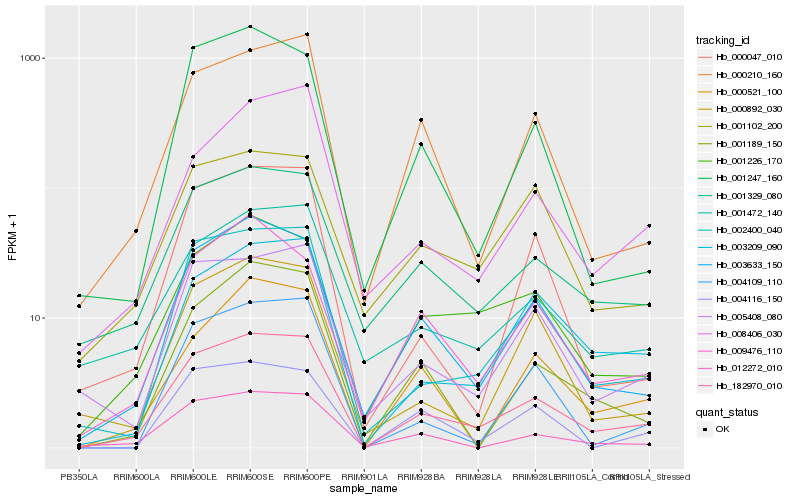
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182970_010 |
0.0 |
- |
- |
calmodulin-binding protein 60-D [Populus trichocarpa] |
| 2 |
Hb_003633_150 |
0.1086614064 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 3 |
Hb_002400_040 |
0.1149391463 |
- |
- |
PREDICTED: uncharacterized protein LOC105637633 [Jatropha curcas] |
| 4 |
Hb_001329_080 |
0.1263101945 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 5 |
Hb_001226_170 |
0.1315670701 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 6 |
Hb_003209_090 |
0.1328326312 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 7 |
Hb_005408_080 |
0.1393663203 |
- |
- |
PREDICTED: U-box domain-containing protein 16 [Jatropha curcas] |
| 8 |
Hb_012272_010 |
0.1408458096 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 9 |
Hb_000210_160 |
0.1412623945 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 10 |
Hb_001472_140 |
0.142267478 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g41330 [Jatropha curcas] |
| 11 |
Hb_001189_150 |
0.1471315596 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 89-like [Jatropha curcas] |
| 12 |
Hb_009476_110 |
0.1502826313 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 13 |
Hb_000521_100 |
0.15212895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001247_160 |
0.1549562996 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 15 |
Hb_000047_010 |
0.1555692895 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 16 |
Hb_008406_030 |
0.1563811762 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 17 |
Hb_004116_150 |
0.1572123938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000892_030 |
0.1581063395 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 19 |
Hb_001102_200 |
0.1585986096 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 20 |
Hb_004109_110 |
0.1593486298 |
- |
- |
dioxygenase family protein [Populus trichocarpa] |