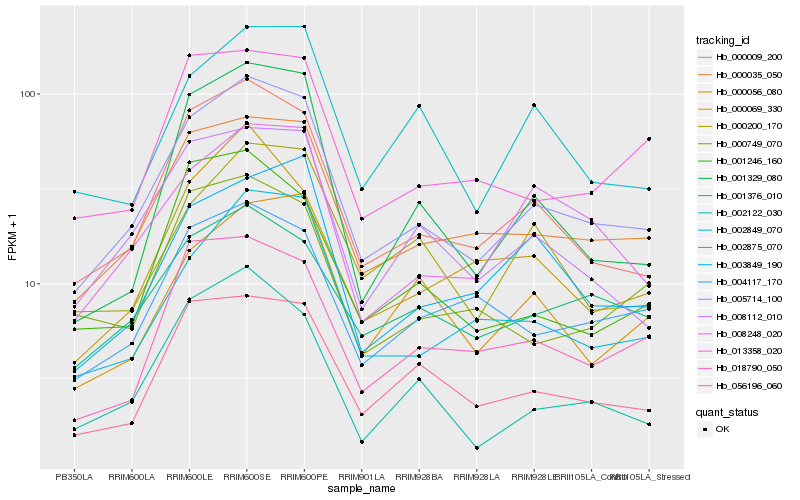
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005714_100 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 2 |
Hb_000009_200 |
0.0735998574 |
- |
- |
PREDICTED: UPF0496 protein At2g18630 [Jatropha curcas] |
| 3 |
Hb_000035_050 |
0.0786790611 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 4 |
Hb_008112_010 |
0.1032567827 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 5 |
Hb_002875_070 |
0.1040811712 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 6 |
Hb_056196_060 |
0.1063390744 |
- |
- |
hexokinase [Manihot esculenta] |
| 7 |
Hb_001376_010 |
0.1129973353 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 8 |
Hb_013358_020 |
0.1157948784 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 9 |
Hb_018790_050 |
0.1165457867 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 10 |
Hb_004117_170 |
0.1168753546 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 11 |
Hb_008248_020 |
0.1174654984 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001329_080 |
0.1191254847 |
- |
- |
PREDICTED: probable protein phosphatase 2C 49 [Jatropha curcas] |
| 13 |
Hb_000069_330 |
0.1212709639 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_002122_030 |
0.1226354054 |
- |
- |
- |
| 15 |
Hb_001246_160 |
0.1229893023 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 16 |
Hb_000749_070 |
0.1255936617 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 17 |
Hb_000056_080 |
0.1270165195 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000200_170 |
0.127124892 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 19 |
Hb_003849_190 |
0.1272726252 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 20 |
Hb_002849_070 |
0.1280250107 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |