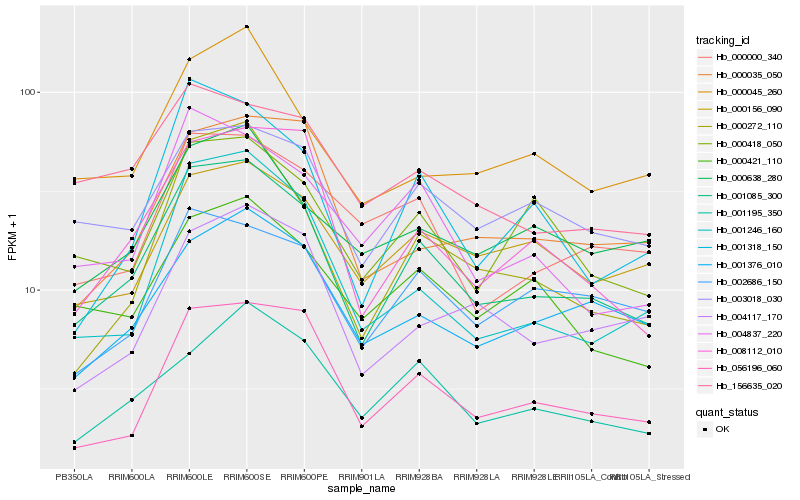
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001085_300 |
0.0 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 2 |
Hb_056196_060 |
0.109430052 |
- |
- |
hexokinase [Manihot esculenta] |
| 3 |
Hb_000272_110 |
0.1102059283 |
- |
- |
PREDICTED: uncharacterized protein LOC105633479 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008112_010 |
0.1134164475 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 5 |
Hb_001318_150 |
0.11513315 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 6 |
Hb_003018_030 |
0.1172547729 |
- |
- |
PREDICTED: GDP-mannose 4,6 dehydratase 1 [Jatropha curcas] |
| 7 |
Hb_004837_220 |
0.1180263466 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 8 |
Hb_001376_010 |
0.118607948 |
- |
- |
diacylglycerol kinase, alpha, putative [Ricinus communis] |
| 9 |
Hb_001195_350 |
0.1187265825 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 10 |
Hb_001246_160 |
0.1191885601 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 11 |
Hb_000421_110 |
0.1216449233 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000418_050 |
0.1220512547 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 13 |
Hb_002686_150 |
0.1252843939 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_000000_340 |
0.1260532413 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 29-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000045_260 |
0.1265179906 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_000156_090 |
0.1267903173 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000638_280 |
0.1269304661 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 18 |
Hb_156635_020 |
0.1284728667 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 19 |
Hb_004117_170 |
0.1295860883 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 20 |
Hb_000035_050 |
0.1307537028 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |