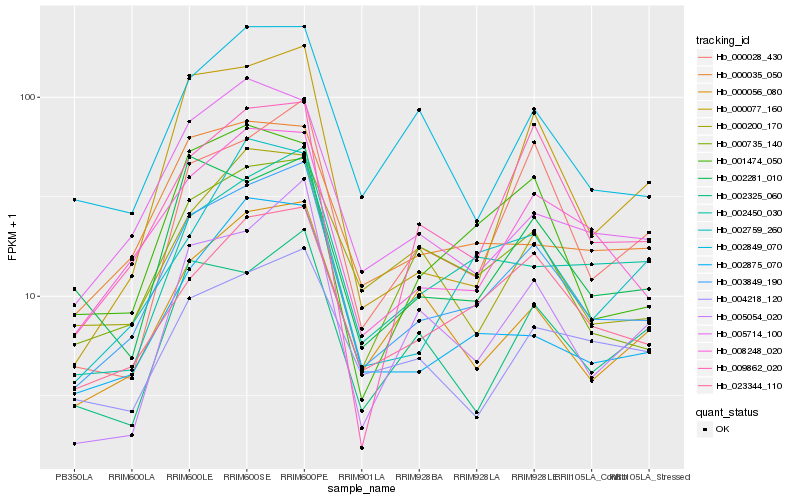
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003849_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 2 |
Hb_008248_020 |
0.0975716005 |
transcription factor |
TF Family: DBB |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_023344_110 |
0.09938331 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 4 |
Hb_000735_140 |
0.1055772759 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 5 |
Hb_002875_070 |
0.1209362625 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 6 |
Hb_002450_030 |
0.1210471123 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 7 |
Hb_000056_080 |
0.1221752085 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000035_050 |
0.1225308618 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 9 |
Hb_005054_020 |
0.1266520365 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 10 |
Hb_000028_430 |
0.1268778816 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005714_100 |
0.1272726252 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 12 |
Hb_002849_070 |
0.1303912026 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001474_050 |
0.1308650268 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000077_160 |
0.1344381702 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 15 |
Hb_002281_010 |
0.1346394843 |
- |
- |
hypothetical protein POPTR_0001s47550g [Populus trichocarpa] |
| 16 |
Hb_000200_170 |
0.1351266953 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Populus euphratica] |
| 17 |
Hb_009862_020 |
0.1354560425 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 18 |
Hb_002759_260 |
0.1379753275 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 19 |
Hb_002325_060 |
0.1386886369 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 20 |
Hb_004218_120 |
0.1393963552 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |