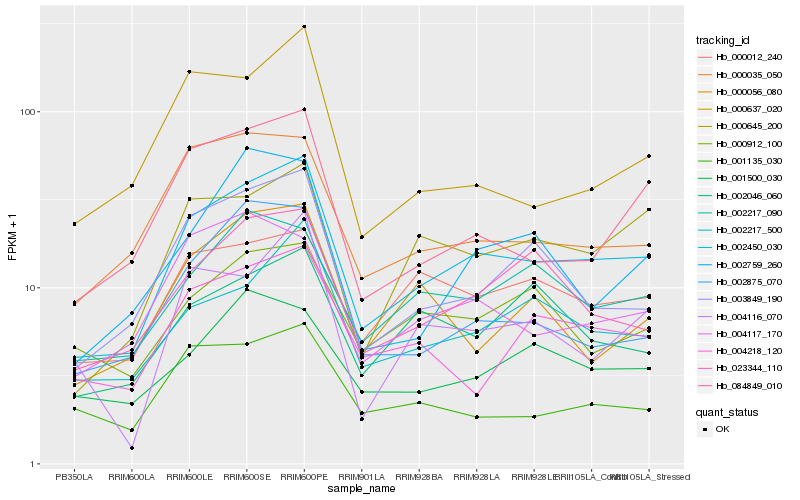
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002450_030 |
0.0 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 2 |
Hb_003849_190 |
0.1210471123 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 3 |
Hb_002217_500 |
0.1245331118 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |
| 4 |
Hb_023344_110 |
0.1300624353 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 5 |
Hb_084849_010 |
0.1320957817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000645_200 |
0.1337351608 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 7 |
Hb_004218_120 |
0.1388493568 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000035_050 |
0.1397787127 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 9 |
Hb_002875_070 |
0.1405513763 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 10 |
Hb_002217_090 |
0.1436627107 |
- |
- |
PREDICTED: probable protein phosphatase 2C 13 [Jatropha curcas] |
| 11 |
Hb_000012_240 |
0.1463786726 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004116_070 |
0.1493892711 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 13 |
Hb_004117_170 |
0.1507983401 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 14 |
Hb_002759_260 |
0.1508195319 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 15 |
Hb_001135_030 |
0.1523390183 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 16 |
Hb_000912_100 |
0.1536625161 |
- |
- |
PREDICTED: CMP-sialic acid transporter 3-like isoform X2 [Elaeis guineensis] |
| 17 |
Hb_000056_080 |
0.1545839645 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000637_020 |
0.1553839222 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 19 |
Hb_002046_060 |
0.1555603021 |
- |
- |
PREDICTED: beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase [Jatropha curcas] |
| 20 |
Hb_001500_030 |
0.1560083006 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |