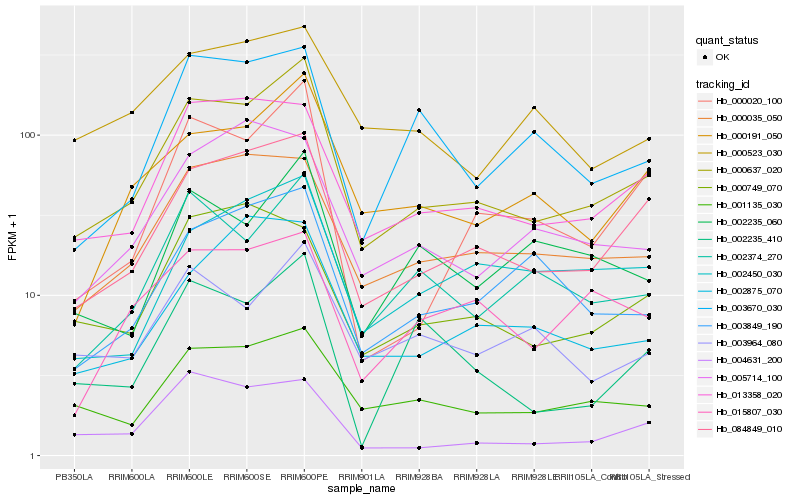
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000637_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650669 [Jatropha curcas] |
| 2 |
Hb_084849_010 |
0.0986500207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001135_030 |
0.1288579559 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 4 |
Hb_000191_050 |
0.1291871193 |
- |
- |
hypothetical protein POPTR_0006s14610g [Populus trichocarpa] |
| 5 |
Hb_013358_020 |
0.1305880447 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 6 |
Hb_000035_050 |
0.1328840274 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 7 |
Hb_002374_270 |
0.1396681939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000020_100 |
0.1407648072 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 9 |
Hb_003849_190 |
0.1542993546 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 10 |
Hb_004631_200 |
0.1551199229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002450_030 |
0.1553839222 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 12 |
Hb_003964_080 |
0.1558635524 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 13 |
Hb_002875_070 |
0.1596097314 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 14 |
Hb_005714_100 |
0.1626208804 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 15 |
Hb_003670_030 |
0.1647034169 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 16 |
Hb_000749_070 |
0.164989667 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 17 |
Hb_002235_060 |
0.1656461236 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 18 |
Hb_002235_410 |
0.1667519535 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 19 |
Hb_000523_030 |
0.1678927868 |
- |
- |
PREDICTED: uncharacterized protein LOC105643073 [Jatropha curcas] |
| 20 |
Hb_015807_030 |
0.1679870609 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |