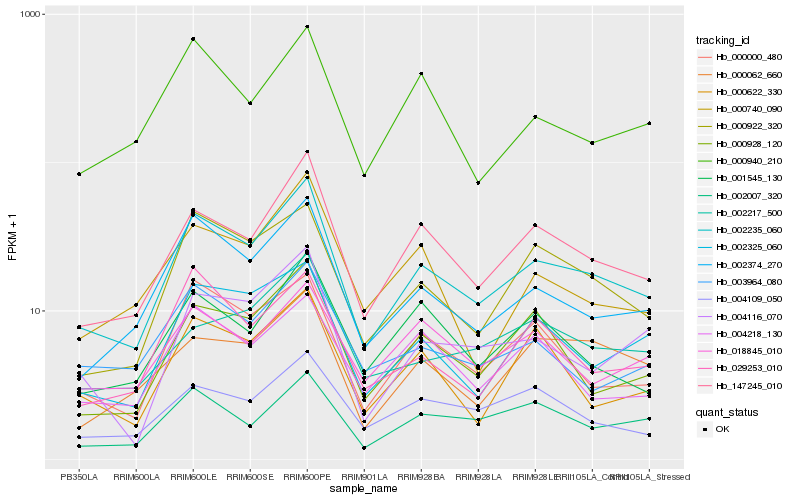
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_060 |
0.0 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_147245_010 |
0.085328979 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 3 |
Hb_002374_270 |
0.0988540803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000922_320 |
0.1132438178 |
- |
- |
hypothetical protein B456_001G071400 [Gossypium raimondii] |
| 5 |
Hb_000000_480 |
0.1159262577 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002325_060 |
0.1181355525 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 7 |
Hb_000928_120 |
0.1226193453 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 8 |
Hb_003964_080 |
0.1306643668 |
- |
- |
PREDICTED: probable methyltransferase PMT5 [Jatropha curcas] |
| 9 |
Hb_001545_130 |
0.1308133497 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 10 |
Hb_004109_050 |
0.1318832442 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 11 |
Hb_004116_070 |
0.1331055787 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 12 |
Hb_029253_010 |
0.1361156475 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 13 |
Hb_000940_210 |
0.1364199613 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 14 |
Hb_002007_320 |
0.136626461 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 15 |
Hb_000740_090 |
0.137665305 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004218_130 |
0.1389170215 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 17 |
Hb_000622_330 |
0.1392849251 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 18 |
Hb_000062_660 |
0.1398714931 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 19 |
Hb_018845_010 |
0.1404719392 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 20 |
Hb_002217_500 |
0.1413246313 |
- |
- |
hypothetical protein JCGZ_19014 [Jatropha curcas] |