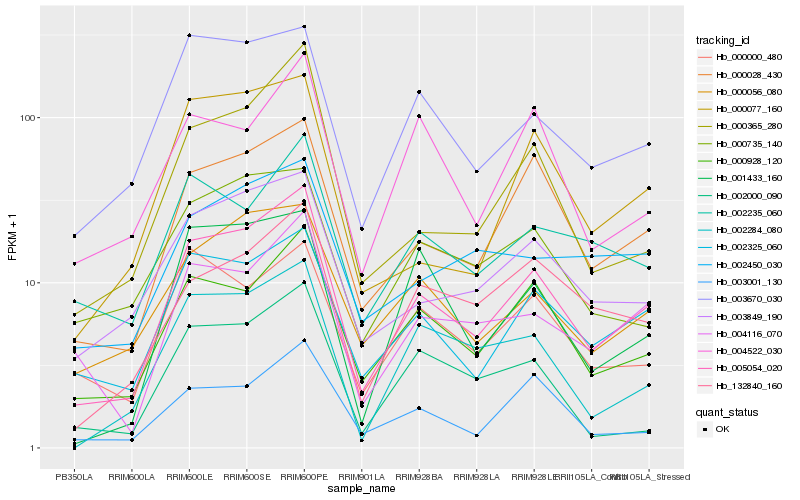
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005054_020 |
0.0 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 2 |
Hb_000028_430 |
0.107564694 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000928_120 |
0.1157948189 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 4 |
Hb_002325_060 |
0.1174997059 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 5 |
Hb_003849_190 |
0.1266520365 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 6 |
Hb_000365_280 |
0.1328701823 |
- |
- |
calcium binding protein/cast, putative [Ricinus communis] |
| 7 |
Hb_000056_080 |
0.1328873468 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003670_030 |
0.1357908681 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 9 |
Hb_004116_070 |
0.1369128587 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 10 |
Hb_003001_130 |
0.1416242201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002284_080 |
0.1437538504 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_000077_160 |
0.1457014628 |
- |
- |
Phospho-N-acetylmuramoyl-pentapeptide-transferase, putative [Theobroma cacao] |
| 13 |
Hb_132840_160 |
0.1463181082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002235_060 |
0.1527494346 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_000000_480 |
0.1567084817 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000735_140 |
0.1582220284 |
- |
- |
3-glucanase family protein [Populus trichocarpa] |
| 17 |
Hb_001433_160 |
0.1586401345 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 18 |
Hb_002450_030 |
0.158845132 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 19 |
Hb_002000_090 |
0.1589684061 |
- |
- |
60S ribosomal protein L3B [Hevea brasiliensis] |
| 20 |
Hb_004522_030 |
0.1599174903 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |