| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
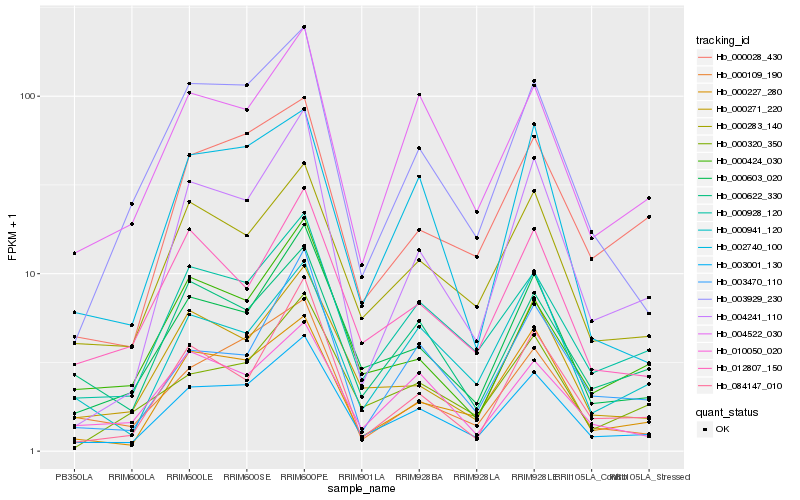
Hb_003001_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000603_020 |
0.0803444083 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000928_120 |
0.0933221703 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 4 |
Hb_000283_140 |
0.1042199084 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004241_110 |
0.1044024273 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |
| 6 |
Hb_000227_280 |
0.1067402434 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 7 |
Hb_000271_220 |
0.108667181 |
- |
- |
PREDICTED: xylosyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_003929_230 |
0.1093581805 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 5 [Nelumbo nucifera] |
| 9 |
Hb_004522_030 |
0.1110869783 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 10 |
Hb_000109_190 |
0.1146360388 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 11 |
Hb_000622_330 |
0.1206549663 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 12 |
Hb_000028_430 |
0.1207852672 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003470_110 |
0.1243409369 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_084147_010 |
0.1252040412 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000320_350 |
0.1257581114 |
- |
- |
hypothetical protein RCOM_0904330 [Ricinus communis] |
| 16 |
Hb_002740_100 |
0.1265405219 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 17 |
Hb_012807_150 |
0.128798244 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 18 |
Hb_010050_020 |
0.1294934477 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 19 |
Hb_000424_030 |
0.1297240902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000941_120 |
0.1299697321 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |