| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
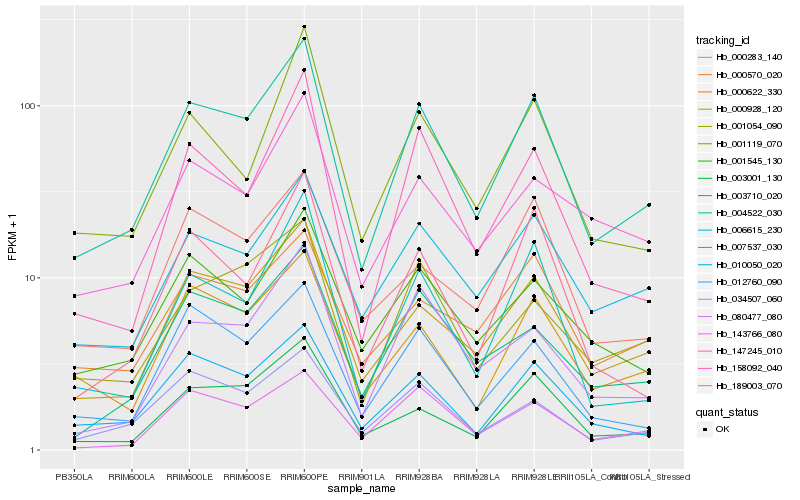
Hb_004522_030 |
0.0 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 2 |
Hb_000928_120 |
0.0805919466 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 3 |
Hb_010050_020 |
0.0995613776 |
- |
- |
PREDICTED: uncharacterized protein LOC102601222 [Solanum tuberosum] |
| 4 |
Hb_007537_030 |
0.1061071673 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000622_330 |
0.1072523115 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 6 |
Hb_003001_130 |
0.1110869783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_034507_060 |
0.1125544984 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic-like [Citrus sinensis] |
| 8 |
Hb_012760_090 |
0.1144761449 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 9 |
Hb_003710_020 |
0.1158084249 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006615_230 |
0.1161471509 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 11 |
Hb_158092_040 |
0.116156559 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 12 |
Hb_001545_130 |
0.117884838 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 13 |
Hb_001054_090 |
0.1183717924 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 14 |
Hb_001119_070 |
0.1201614775 |
- |
- |
methylenetetrahydrofolate reductase, putative [Ricinus communis] |
| 15 |
Hb_143766_080 |
0.1201689943 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000283_140 |
0.1217833988 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_147245_010 |
0.123893918 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 18 |
Hb_189003_070 |
0.1241188455 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 19 |
Hb_080477_080 |
0.1246747451 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 20 |
Hb_000570_020 |
0.1248314319 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |