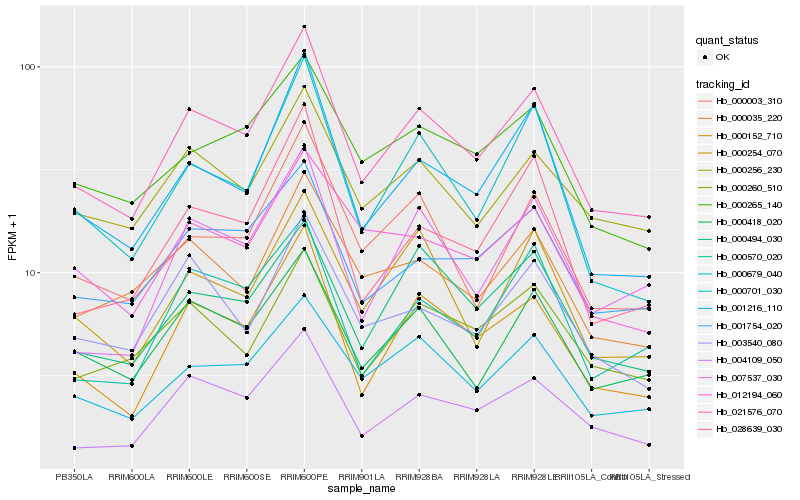
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021576_070 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 2 |
Hb_000152_710 |
0.0660960938 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 3 |
Hb_004109_050 |
0.0744264653 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 4 |
Hb_000701_030 |
0.0770515439 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 5 |
Hb_001216_110 |
0.079708263 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 6 |
Hb_000418_020 |
0.0814287119 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001754_020 |
0.0816075845 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 8 |
Hb_000256_230 |
0.0870412576 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 9 |
Hb_000003_310 |
0.0877699723 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 10 |
Hb_000494_030 |
0.0881243382 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 11 |
Hb_000679_040 |
0.0885726911 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 12 |
Hb_000265_140 |
0.0897202851 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 13 |
Hb_000035_220 |
0.0910929642 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 14 |
Hb_000260_510 |
0.0919857872 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 15 |
Hb_000254_070 |
0.09402218 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 16 |
Hb_003540_080 |
0.0956643993 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_007537_030 |
0.0964126995 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_028639_030 |
0.0968154919 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 19 |
Hb_012194_060 |
0.0979205856 |
- |
- |
PREDICTED: uncharacterized protein LOC105636108 [Jatropha curcas] |
| 20 |
Hb_000570_020 |
0.0979765519 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |