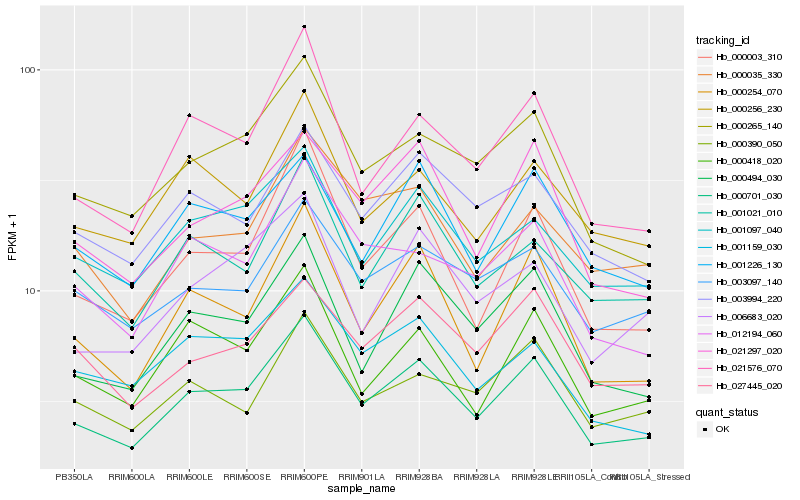
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000701_030 |
0.0 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 2 |
Hb_000265_140 |
0.0707019241 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 3 |
Hb_021576_070 |
0.0770515439 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 4 |
Hb_000494_030 |
0.0779698188 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 5 |
Hb_000254_070 |
0.0788233439 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 6 |
Hb_000003_310 |
0.0812963708 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 7 |
Hb_001097_040 |
0.0814014479 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 8 |
Hb_003994_220 |
0.0836456776 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 9 |
Hb_021297_020 |
0.0846726959 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 10 |
Hb_001021_010 |
0.0874522626 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 11 |
Hb_000418_020 |
0.0878036866 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001226_130 |
0.0884963016 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 13 |
Hb_000256_230 |
0.0902538331 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 14 |
Hb_003097_140 |
0.0903750759 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 15 |
Hb_027445_020 |
0.0907089779 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 16 |
Hb_012194_060 |
0.0907458488 |
- |
- |
PREDICTED: uncharacterized protein LOC105636108 [Jatropha curcas] |
| 17 |
Hb_001159_030 |
0.0933073817 |
- |
- |
PREDICTED: probable protein S-acyltransferase 14 [Jatropha curcas] |
| 18 |
Hb_000035_330 |
0.0947641396 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 19 |
Hb_000390_050 |
0.0956498445 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 20 |
Hb_006683_020 |
0.0965250401 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |