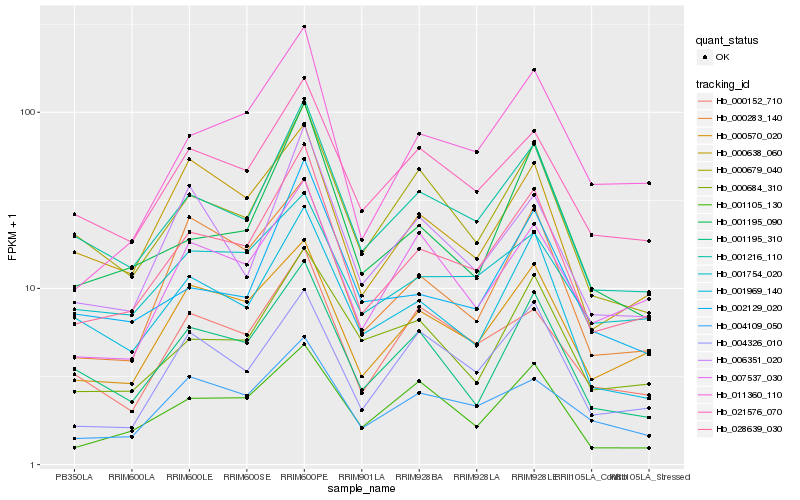
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028639_030 |
0.0 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 2 |
Hb_001216_110 |
0.0731933272 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 3 |
Hb_000570_020 |
0.0923242431 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 4 |
Hb_021576_070 |
0.0968154919 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 5 |
Hb_011360_110 |
0.1014260254 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 6 |
Hb_000679_040 |
0.104508146 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 7 |
Hb_001195_090 |
0.1073529468 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 8 |
Hb_001969_140 |
0.109843788 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 9 |
Hb_001195_310 |
0.1099524845 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_004109_050 |
0.1102414832 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 11 |
Hb_000283_140 |
0.1106836124 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_002129_020 |
0.1126109546 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 13 |
Hb_001105_130 |
0.1139759431 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 14 |
Hb_000152_710 |
0.1141429074 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 15 |
Hb_000684_310 |
0.1142294247 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 16 |
Hb_006351_020 |
0.1163112304 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 17 |
Hb_007537_030 |
0.1163947904 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000638_060 |
0.1169130906 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |
| 19 |
Hb_004326_010 |
0.1191647338 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_001754_020 |
0.1196473723 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |