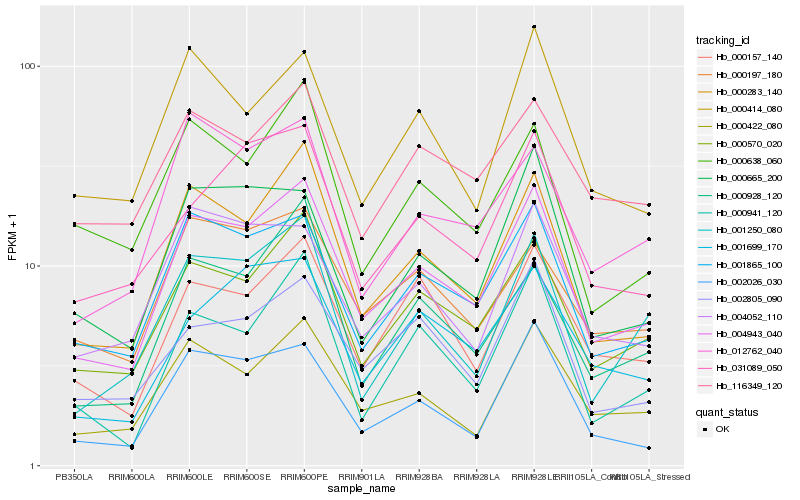
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004943_040 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000570_020 |
0.07124931 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 3 |
Hb_000283_140 |
0.0727126501 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001865_100 |
0.0790052749 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 5 |
Hb_000422_080 |
0.0911023187 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 6 |
Hb_000157_140 |
0.1010637348 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_002805_090 |
0.107520574 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_002026_030 |
0.108843024 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 9 |
Hb_031089_050 |
0.1125040204 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_000665_200 |
0.1130204983 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_001250_080 |
0.1134310947 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g23390 [Jatropha curcas] |
| 12 |
Hb_000414_080 |
0.1146867063 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000941_120 |
0.1149859123 |
- |
- |
hypothetical protein JCGZ_25565 [Jatropha curcas] |
| 14 |
Hb_000197_180 |
0.1150411038 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 15 |
Hb_004052_110 |
0.1153842644 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000928_120 |
0.1165915633 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 17 |
Hb_116349_120 |
0.1174300038 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_012762_040 |
0.1176302913 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 19 |
Hb_001699_170 |
0.1177001529 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000638_060 |
0.1197657769 |
- |
- |
PREDICTED: probable methyltransferase PMT26 [Jatropha curcas] |