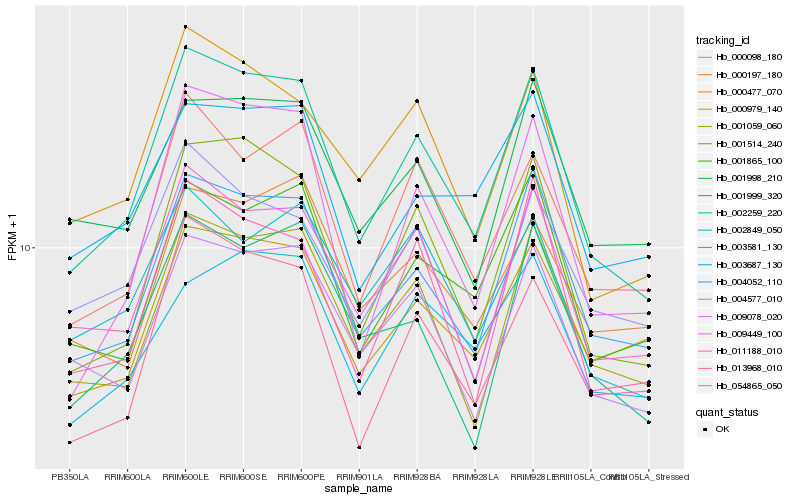
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_220 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 2 |
Hb_004052_110 |
0.0665305523 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001514_240 |
0.0750378349 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 4 |
Hb_011188_010 |
0.0833290091 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009078_020 |
0.0903518623 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 6 |
Hb_001059_060 |
0.0915221213 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 7 |
Hb_000477_070 |
0.0931516505 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 8 |
Hb_000979_140 |
0.0962420031 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 9 |
Hb_001865_100 |
0.0964224692 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 10 |
Hb_009449_100 |
0.0968825921 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 11 |
Hb_000098_180 |
0.0994892031 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 12 |
Hb_013968_010 |
0.1011791709 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 13 |
Hb_001999_320 |
0.1075310321 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 14 |
Hb_001998_210 |
0.1084786377 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_003687_130 |
0.1117178142 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 16 |
Hb_054865_050 |
0.1119845903 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 17 |
Hb_002849_050 |
0.112115763 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 18 |
Hb_004577_010 |
0.1147588565 |
- |
- |
PREDICTED: transmembrane protein 45A [Prunus mume] |
| 19 |
Hb_003581_130 |
0.1168558319 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000197_180 |
0.1174869907 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |