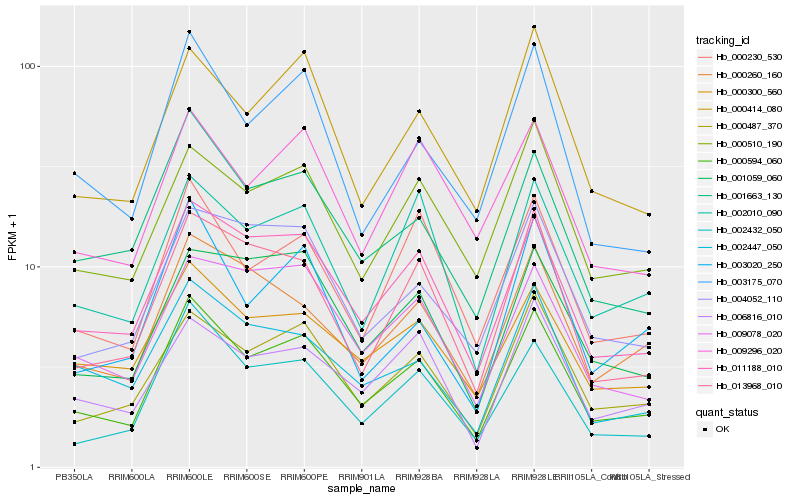
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000594_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 2 |
Hb_011188_010 |
0.0768261307 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000487_370 |
0.0835849961 |
- |
- |
- |
| 4 |
Hb_002432_050 |
0.0899739483 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 5 |
Hb_001663_130 |
0.0915109321 |
- |
- |
acyl-CoA thioesterase, putative [Ricinus communis] |
| 6 |
Hb_003175_070 |
0.0947491063 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 7 |
Hb_000414_080 |
0.0961824261 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_013968_010 |
0.1003609169 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 9 |
Hb_000230_530 |
0.1015242789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000300_560 |
0.1017062974 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000510_190 |
0.1032703884 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 12 |
Hb_002010_090 |
0.104442862 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000260_160 |
0.1055756664 |
transcription factor |
TF Family: GNAT |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_001059_060 |
0.1065189428 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 15 |
Hb_002447_050 |
0.1066118392 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_006816_010 |
0.1069513859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004052_110 |
0.1077371957 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_009078_020 |
0.1089926054 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 19 |
Hb_003020_250 |
0.1094602495 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 20 |
Hb_009296_020 |
0.1133570243 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |