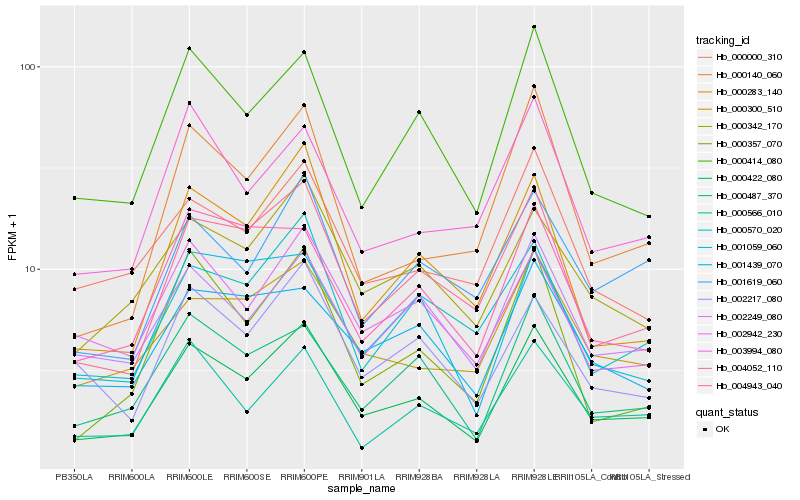
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000422_080 |
0.0 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 2 |
Hb_004943_040 |
0.0911023187 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000414_080 |
0.0928964066 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000487_370 |
0.0989910854 |
- |
- |
- |
| 5 |
Hb_000342_170 |
0.1003921055 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 6 |
Hb_000283_140 |
0.1022432878 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002249_080 |
0.1063620501 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 8 |
Hb_000300_510 |
0.1105458687 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 9 |
Hb_000570_020 |
0.1119183725 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 10 |
Hb_000140_060 |
0.1120510808 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 11 |
Hb_002942_230 |
0.1136990854 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 12 |
Hb_000566_010 |
0.1150110312 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 13 |
Hb_000000_310 |
0.1152486705 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 14 |
Hb_002217_080 |
0.1164371741 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 16 [Jatropha curcas] |
| 15 |
Hb_003994_080 |
0.1174457032 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 16 |
Hb_001619_060 |
0.1178248417 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001439_070 |
0.1195677984 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000357_070 |
0.1199752486 |
- |
- |
- |
| 19 |
Hb_004052_110 |
0.1201265464 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001059_060 |
0.1218250248 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |